Figure 3.
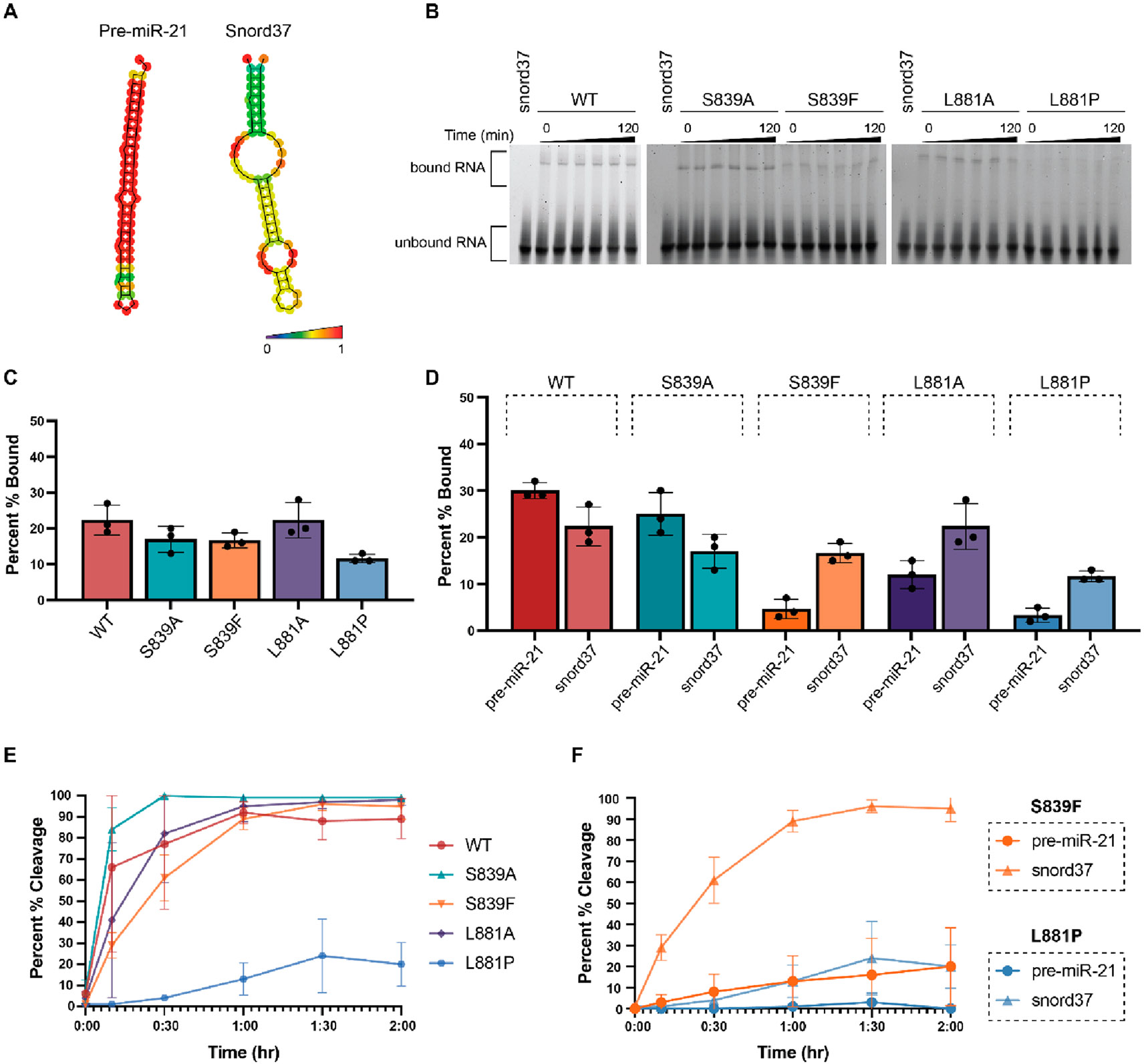
Characterization of the binding and cleavage activity of WT and Dicer mutants for snord37. (A) Predicted secondary structures of pre-miR-21 and snord37 as determined via RNAfold 2.4.18. The color scale indicates base-pair probabilities. (B) Representative EMSA data from three independent experiments (Figure S3) shown over the course of 120 min. (C) Quantification of binding at the 120 min time point as determined using the relative difference in intensity of bound RNA (top band) to unbound RNA (bottom band). (D) Comparison of binding to pre-miR-21 and snord37 at 120 min. (E) Quantification of time-dependent Dicer cleavage activity (Figure S4). Gel-based analysis was carried out using ImageJ. Error bars represent the standard deviation from three independent experiments (Tables S1 and S2). (F) Comparison of cleavage of pre-miR-21 and snord37 at 120 min by Dicer disease mutants S839F and L881P.
