Abstract
The RNase III endoribonuclease Dicer was discovered to be associated with cleavage of double-stranded RNA in 2001. Since then, many advances in our understanding of Dicer function have revealed that the enzyme plays a major role not only in microRNA biology but also in multiple RNA interference-related pathways. Yet, there is still much to be learned regarding Dicer structure–function in relation to how Dicer and Dicer-like enzymes initiate their cleavage reaction and release the desired RNA product. This Perspective describes the latest advances in Dicer structural studies, expands on what we have learned from this data, and outlines key gaps in knowledge that remain to be addressed. More specifically, we focus on human Dicer and highlight the intermediate processing steps where there is a lack of structural data to understand how the enzyme traverses from pre-cleavage to cleavage-competent states. Understanding these details is necessary to model Dicer’s function as well as develop more specific microRNA-targeted therapeutics for the treatment of human diseases.
Graphical Abstract
MicroRNAs (miRNA) are a family of single-stranded, noncoding RNAs involved in regulating protein-coding genes via post-transcriptional silencing of mRNA translation.1–6 The multistep, protein-mediated process of the miRNA biogenesis pathway is critical in ensuring that proper miRNA expression levels and seed sequence integrity are maintained.4,5,7 The canonical miRNA biogenesis pathway consists of sequential cleavage reactions performed by the RNase III enzymes, Drosha and Dicer. Noncanonical pathways are also present in which miRNA processing is Drosha or Dicer independent, but this is less common.8–10 Alterations to these pathways, causing either the overexpression or loss of miRNAs, are linked to a variety of human diseases, including cancers.11,12 Attempts to regulate this activity via directly targeting miRNAs remain challenging due to difficulties associated with the development of RNA-based miRNA inhibitors and mimetics, as well as the selective targeting of miRNAs with small molecules.12–16 With respect to the latter, a deeper functional and structural understanding of the proteins involved in the miRNA biogenesis pathway and, more specifically, how these proteins interact with their RNA substrates is required to aid inhibitor design and development.
The canonical miRNA biogenesis pathway begins in the nucleus where a miRNA gene is transcribed by RNA polymerase II (Figure 1).5,17–19 The resulting transcript folds back on itself to form the hairpin loop structure of a primary miRNA (pri-miRNA) with long overhang regions at the flanking 5′ and 3′ ends. Pri-miRNAs then undergo two processing steps taking place in the nucleus and cytoplasm by the RNase III-type enzymes Drosha and Dicer, respectively. During the first cleavage reaction, Drosha and its cofactor DiGeorge Syndrome Critical Region 8 (DGCR8) cleave off the flanking regions of the pri-miRNA, excising out the precursor miRNA (pre-miRNA) hairpin loop. The pre-miRNA is then exported to the cytoplasm by Exportin 5 for the final cleavage reaction performed by Dicer and its cofactor, Transactivation Response Element RNA-Binding Protein (TRBP). Dicer cleaves off the loop region of the pre-miRNA to form the mature miRNA duplex consisting of the 5 prime (5P) and 3 prime (3P) strands.4,17 Either the 5P or 3P strand from the mature miRNA duplex is then loaded into Argonaute 2 (Ago2) to form the RNA-induced silencing complex (RISC).20,21 One proposed mechanism of strand selection between Ago2 and human Dicer suggests that it is based on the thermodynamic stability of the 5′ end of the miRNA duplex; however, it is still not fully understood if additional factors dictate this process or if Dicer proteins from varying species perform an altogether alternative mechanism depending on the RNA substrate.20,22–26 After RISC formation, this complex targets mRNAs based on complementarity to the miRNA guide strand sequence to mark the mRNA for translational repression and/or degradation (Figure 1).1,3,7,27–29
Figure 1.
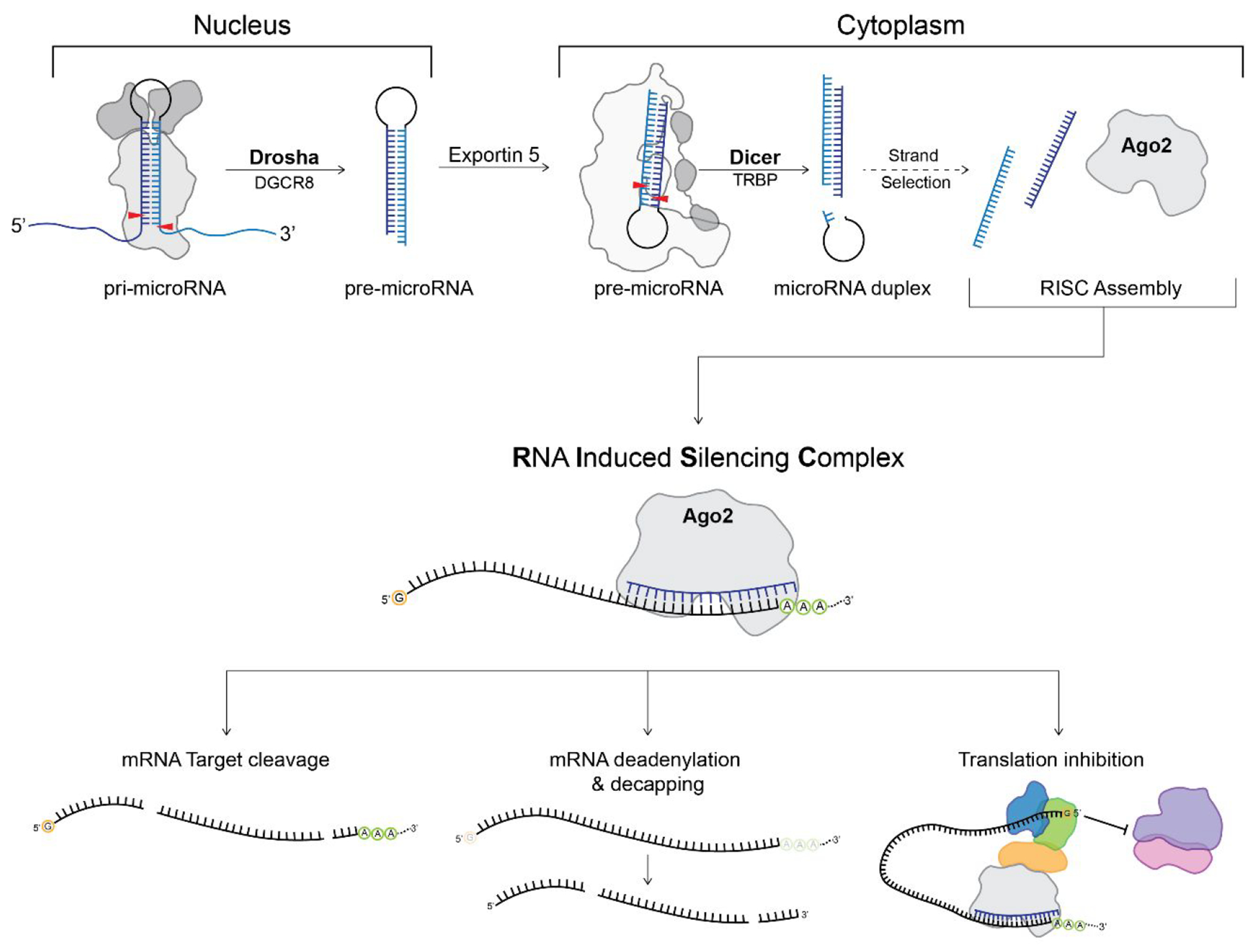
Canonical miRNA biogenesis pathway highlighting the processing steps carried out by Drosha and Dicer. The 5P and 3P strands of microRNA are shown in dark blue and light blue, respectively, while the red arrows indicate the sites of cleavage. Purple and pink represent the 50S and 30S ribosome with additional proteins recruited for translation inhibition by RISC shown in orange, royal blue, and green. The 7-methlyguanosine 5′ mRNA cap is represented by an orange circle with the letter “G”, while the 3′ poly(A) tail is shown as a green circle with the letter “A”.
STRUCTURAL DIVERSITY AMONG RNase III FAMILY ENZYMES
Despite performing similar cleavage reactions in the miRNA biogenesis pathway, Drosha and Dicer fall into separate classes among the RNase III family of enzymes.30 These differences are related to the non-RNase III domains that ultimately diversify the primary functions and substrate specificity of each class within the RNase III family. Figure 2 shows an overview of the domain organization of RNase III enzymes found in each class and how their domain distributions compare across species.17,26,31,32
Figure 2.
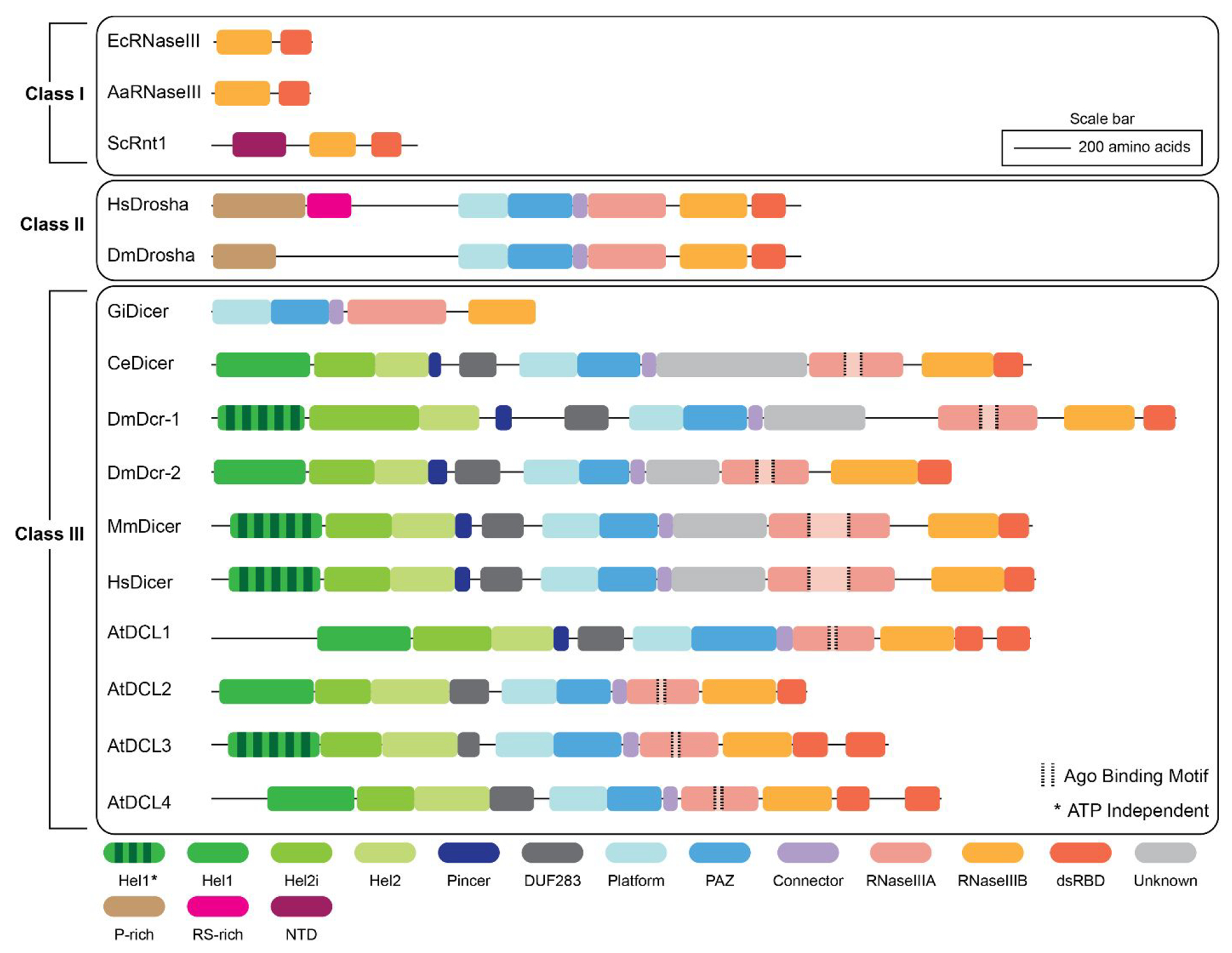
Domain organization and distribution of the Class I, II, and III RNase III family of enzymes. Ec – E. coli; Aa – A. aeolicus; Sc – S. cerevisiae; Hs – Human; Dm – Drosophila; Gi – Giardia; Ce – C. elegans; At – Arabidopsis.
Class I RNase III enzymes are the smallest family members ranging between ~200 and 300 amino acids in length, and accordingly, based on their limited complexity, are the most well-characterized of the classes.30,33 Class I RNase III enzymes are found ubiquitously in bacteria, bacteriophage, and some fungi and contain a single RNase III domain and a double-stranded RNA-binding domain (dsRBD).30 These enzymes cleave double-stranded RNA (dsRNA) substrates through the homodimerization of two RNase III domains to form a single processing center. The overall structure of this homodimeric RNase III processing center is highly conserved across all Class I RNase III enzymes;26 however, in higher eukaryotes that rely on Class II and/or Class III RNase III enzymes, the processing center is composed of two separate RNase III domains, RNase IIIA and RNase IIIB.34–37 This evolutionary change from a simple homodimeric network to a more complex heterodimeric internal processing center suggests the need for higher eukaryotes to have added control in differentiating their cleavage substrates and performing more precise roles in RNA interference (RNAi) pathways. In line with this hypothesis, Class II and Class III enzymes also contain a larger variety of noncatalytic domains that function in substrate binding and recognition.
Drosha enzymes, representing the Class II RNase III family, contain a heterodimeric RNase III processing center, as well as a PAZ-like and Platform domain within the central domain of Drosha (CED).38,39 Proline-rich (P-rich) and arginine serine-rich (RS-rich) domains are also present in most Drosha species and may serve to localize the enzyme to the nucleus.38,40 Drosha works in tandem with its cofactor DGCR8 to bind to pri-miRNA substrates and perform its cleavage reaction. Unlike Dicer’s interaction with TRBP, which has been shown to be dispensable for cleavage activity, Drosha is unable to cleave primiRNAs in the absence of DGCR8. The PAZ-like and Platform domains in Drosha function in binding to the stem region near the cut site of the pri-miRNA, while two DGCR8 proteins coordinate to bind the hairpin loop region.38,39 The cleavage reaction performed by Drosha leaves the characteristic 3′ 2 nucleotide (nt) overhang that is needed for Dicer to recognize and bind the resulting pre-miRNA substrate.41,42
The Class III RNase III family is composed of Dicer and Dicer-like enzymes. The defining structural difference between Class II and Class III proteins is the addition of an N-terminal duplex RNA-activated ATPase (DRA) helicase domain, also sometimes referred to as a DExD/H-box helicase domain.43–46 The DRA helicase domain, formed by three subdomains, Hel1, Hel2i, and Hel2, binds and unwinds the hairpin loop region of the pre-miRNA substrate.44,47–49 As shown in Figure 2, while the majority of Dicer and Dicer-like enzymes contain an N-terminal DRA helicase domain, there is variability between species regarding whether this domain is ATP dependent or independent. These differences in ATP dependence are seemingly linked to Dicer’s substrate specificity during cleavage.44,49–51 For instance, in Drosophila, Dicer-1 is ATP independent and primarily cleaves pre-miRNA, while Dicer-2 requires ATP to cleave pre-small interfering RNAs (siRNA).49,51–54 Interestingly, the minimal Giardia Dicer lacks the helicase domain altogether despite being classified as a Class III RNase III enzyme.55 Lack of a helicase domain may suggest an evolutionary shift in Dicer enzymes requiring a more sophisticated means to differentiate RNA substrates in higher eukaryotes. Therefore, variability in ATP dependence and/or the presence of a helicase domain may regulate the types of RNA substrates that Dicer can recognize and cleave; however, it is currently unclear how or why certain Dicer species have evolved these divergent mechanisms.44,56
The RNase IIIA domain of Dicer also contains a degree of sequence diversity within the Class III RNase III family. In human and mouse Dicers, a large sequence insertion adds 200 amino acids in the middle of the RNase IIIA domain and binds to Ago2.57,58 This stretch of amino acids is referred to as the Ago2 minimal binding motif and is unique to the Class III RNase III enzymes. When the insertion in the RNase IIIA domain is deleted, a decrease in Ago2 binding to human Dicer (hsDicer) is observed, suggesting that this region may by a critical binding point for Ago2 loading.58 While there are currently no structures of this motif/region for hsDicer, recent structures of Drosophila Dicer-2 show that this motif may facilitate dmDicer-2 dimerization.54 This recent discovery raises questions of how Dicer proteins in lower eukaryotes, such as Arabidopsis and Drosophila, utilize this sequence insertion, perhaps for alternative functions outside of Ago2 binding and RISC loading. Further investigation is needed to determine if this binding motif plays a role in RISC assembly during the strand selection process when Ago2 is present and if this mechanism is used across Dicer-containing species. One hypothesis we propose, which would be specific to humans, is that Dicer acts as a scaffolding unit for Ago2 to bind and load the mature miRNA guide strand in contrast to Dicer simply releasing the mature miRNA duplex or a handoff process occurring through TRBP alone.59
The dynamic and multifunctional nature of Dicer has historically made the functional and structural characterization of this enzyme challenging; however, the resolution revolution in cryo-electron microscopy (cryo-EM) has led to structures of Dicer from a variety of species at resolutions conducive to building detailed molecular models that can then be tested biochemically or in vivo.60–62 Yet, even with these breakthroughs, mechanistic questions remain. For example, the lack of structural information about human Dicer in cleavage-competent and post-cleavage states is still required for interpreting how human Dicer’s noncatalytic domains function during the cleavage reaction. Additionally, it is unclear how sequence diversity among Dicer proteins in various species alters structure and function. Until additional hsDicer structures are determined at various stages of RNA processing, we will continue to struggle to accurately model interactions with RNA substates before, during, and after the cleavage reaction. In this Perspective, we discuss what is known about Dicer function and structure; summarize what mechanistic information regarding human Dicer remains to be discovered; present a hypothetical model for the initiation, cleavage, and release of miRNA using currently available data; and provide insights into future studies necessary to answer fundamental questions regarding the role of Dicer structure–function in miRNA processing and biology.
ADVANCES IN DICER STRUCTURAL STUDIES
Since the discovery of Dicer in 2001,63 our biochemical understanding of this Class III RNase III enzyme has quickly developed, while structural information was at first slow to advance. This is due to the size, complexity, and flexibility of Class III enzymes making structural studies of full-length Dicer and Dicer-like enzymes using traditional structural techniques such as X-ray crystallography and NMR challenging.60,62 However, during the resolution revolution of cryo-EM, protein and protein complexes previously thought of as unsolvable became attainable.60,61,64,65 Figure 3 shows a snapshot of major structural breakthroughs between 2001 and 2022 that have aided our understanding of Dicer structure and function.
Figure 3.
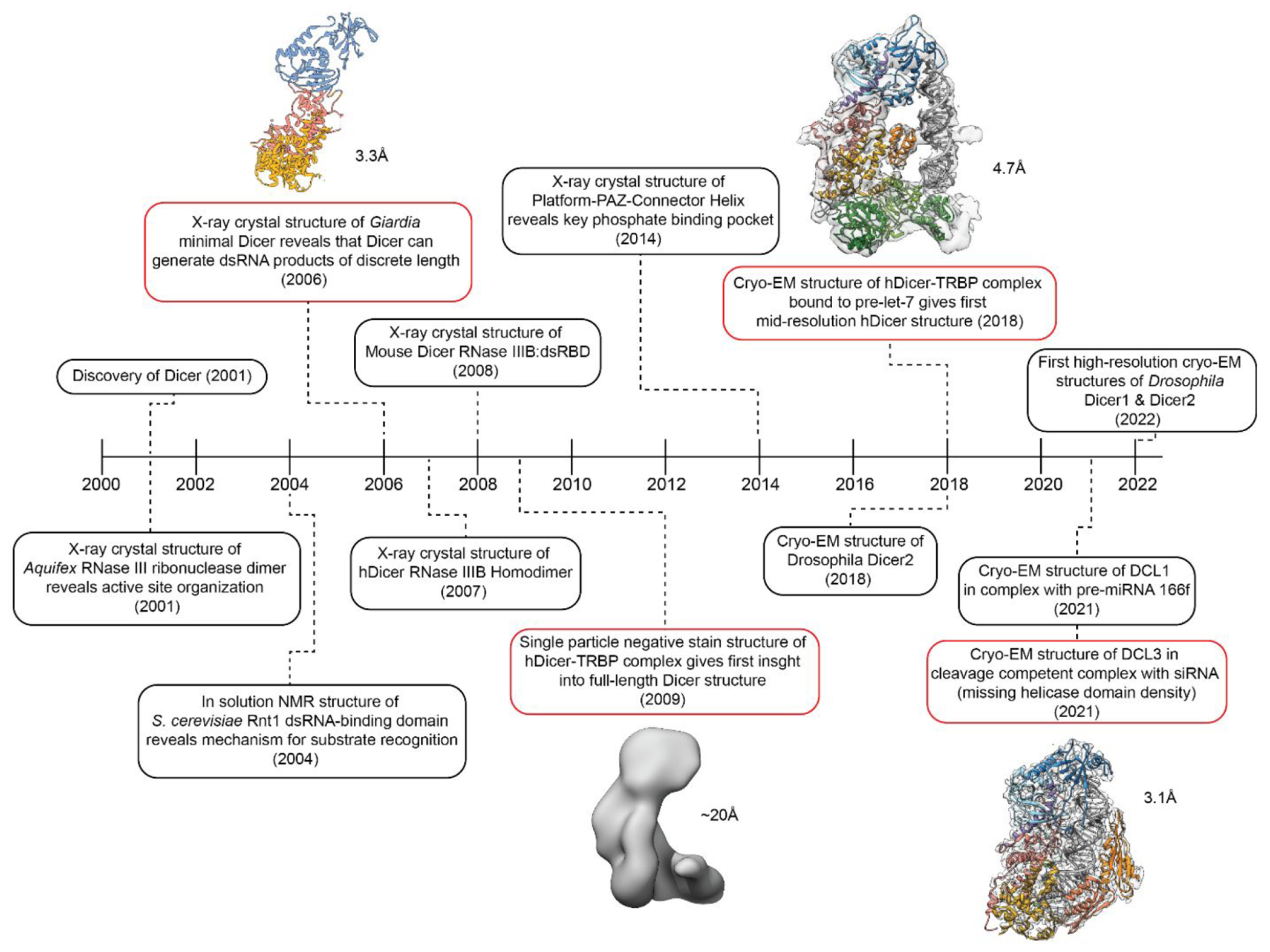
Timeline of major findings that have expanded the understanding of the structural organization of Dicer. Significant advances that contributed to new information regarding full-length Dicer and Dicer-like structures are shown in red with their corresponding resolutions.
Initial structural studies of Dicer and related RNase III enzymes first relied on the analysis of the simpler Class I RNase IIII enzymes. In 2001, X-ray crystallographic studies of the Aquifex RNase III domain provided the first look at active site organization and the characteristic dimerization pattern of the RNase III processing center which is conserved across all RNase III family enzymes.35–37 Shortly after, NMR structural studies of S. cerevisiae Rnt1 RNase III revealed a mechanism for substrate recognition that is dependent on RNA structure and illustrated how accessory domains contribute to substrate specificity.66 Although these first structures introduced the field to key attributes of RNase III enzymes, such as active site dimerization and RNA binding patterns, they only allowed for conjecture about the mechanisms used by the multidomain Class II and Class III RNase III enzymes. Additionally, these early studies provided no structural information on how or why Class III RNase III enzymes utilize a helicase domain to perform their downstream cleavage reactions.
To expand upon these initial structures and begin addressing key mechanistic questions related to Class II and Class III enzymes, researchers first focused on understanding how Dicer performs its cleavage reaction with product-length specificity. Dicer’s ability to cleave dsRNA to a particular length is one of its defining functions compared to Class I RNase III enzymes.19,36,55 To answer this question, structural studies of more complex and larger Dicer species were required, but during this early period (pre-2013), the only structural techniques available that would also yield a resolution quality high enough to build the molecular models needed to answer detailed mechanistic questions were limited by protein size and flexibility.60,62 As such, researchers focused on the investigation of the minimal Dicer from Giardia and determining structures of individual Dicer domains. Due to its smaller size and lack of flexible helicase domain, Giardia Dicer enabled characterization of the functional core of Dicer (Platform-PAZ and RNase III) using X-ray crystallography.55 This structure showed that not only was the PAZ domain responsible for binding to the 3′ 2 nucleotide (nt) overhang of dsRNA but also a 65 Å distance between the PAZ and RNase III processing center matched the length of mature miRNA products of ~22–25 nt. This finding made it possible to propose a molecular model for how Dicer can precisely cleave dsRNA to a predetermined length in which the physical distance between the PAZ RNA-binding pocket and the catalytic residues in the RNase III domains act as a ruler for cleaving dsRNA. Nearly 8 years after this discovery, structural characterization of the Platform-PAZ-Connector domain using X-ray crystallography also revealed a 5′ phosphate binding pocket contained within the Platform domain adding additional insight into the Dicer cleavage mechanism.67 However, the lack of the helicase domain in Giardia Dicer and structural studies of only Dicer’s Platform-PAZ and RNase III domains made it difficult to understand how this essential helicase domain in hsDicer or Dicer-like species of higher complexity contributes to the cleavage of dsRNA.
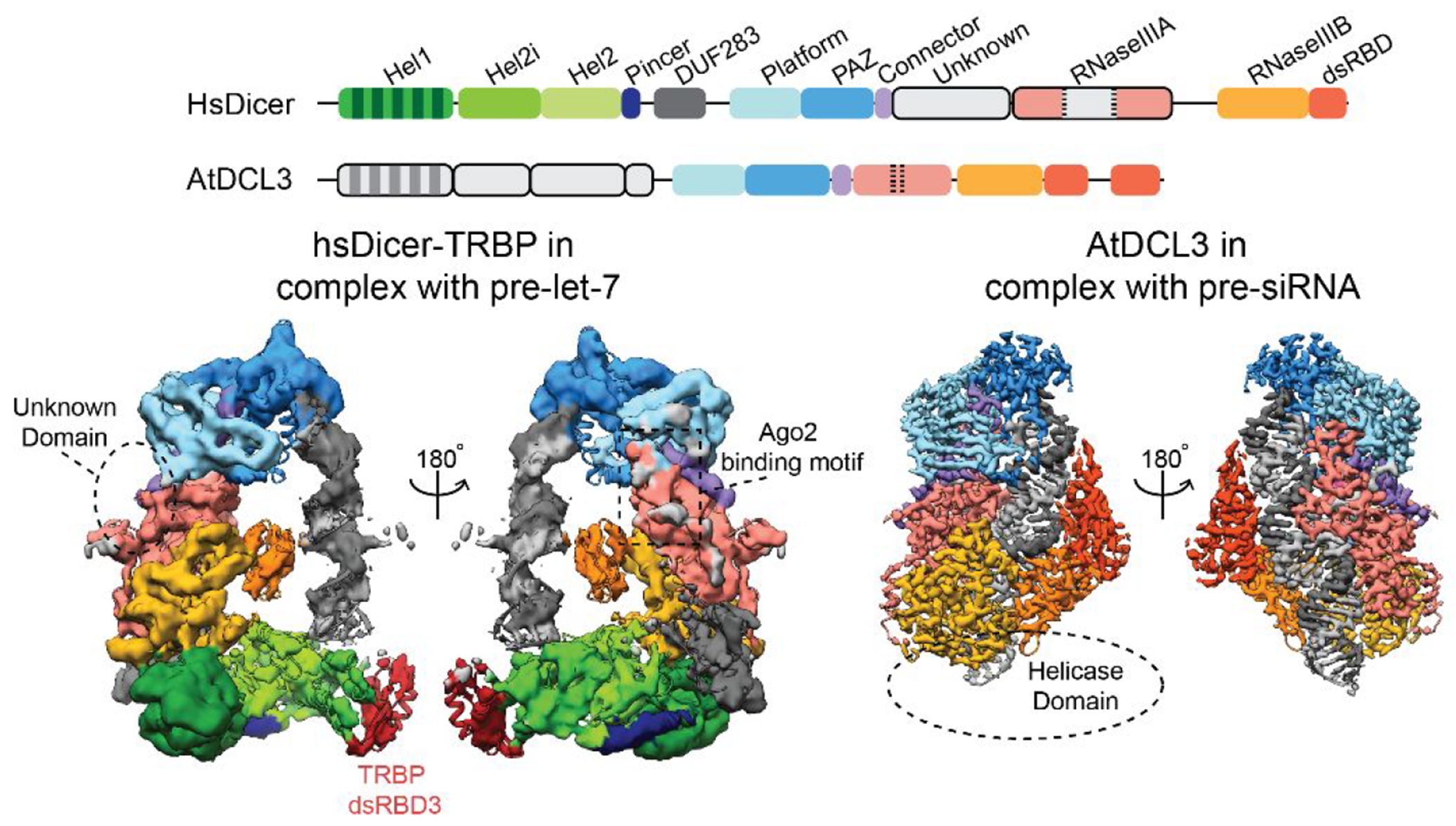
Early attempts at studying full-length hsDicer to better characterize the helicase domain and additional noncatalytic domains were done in 2009 and 2012 using negative stain transmission electron microscopy (TEM).68,69 Although only reaching ~20 Å resolution, the structures showed that Dicer forms a L-shaped structure and provided an initial model for the structural organization of hsDicer. Later advancements in the field of cryo-EM, both instrumentation and computation, made it possible to determine structures of full-length Dicer from humans, Drosophila, and Arabidopsis at resolutions amenable for building molecular models despite its highly flexible nature.52–54,60,70,71 As a result, by the end of 2021, the first four full-length Dicer and Dicer-like structures were published with resolutions spanning 7 to 3.1 Å. These include structures of Drosophila Dicer-2,49 Apo-hsDicer-TRBP complex,70 hsDicer-TRBP bound to pre-let-7 in a pre-cleavage state,70 and a cleavage-competent structure of Arabidopsis Dicer-like-protein 3 (DCL3) bound to pre-siRNA.71 In addition to these structures, new structures continue to be reported including six Drosophila Dicer-1–Loquacious-PB (Loqs-PB) complex structures,52 six Drosophila Dicer-2–Loqs-PD complex54 structures, and two Drosophila Dicer-2–R2D2 complex structures.53 The 14 Drosophila Dicer structures depict dmDicer-1 and dmDicer-2 both apo- and in complex with dsRNA spanning a variety of conformational states. This quickly growing pool of structural knowledge has provided insight into the Drosophila Dicer mechanism, which is described later; however, in terms of human Dicer, our structural knowledge remains lagging.
CRYO-EM STUDIES OF DROSOPHILA DICER-1 AND DICER-2 REVEAL STEPWISE MECHANISM
Unlike humans which only contain one copy of Dicer, Drosophila has evolved two copies of Dicer known as Dicer-1 and Dicer-2.26,43 As previously mentioned, these Dicer proteins differ in their ATP dependence with dmDicer-1 cleaving pre-miR substrates independent of ATP, while dmDicer-2 depends on ATP to cleave long dsRNA and discriminate between the dsRNA termini for specific cleavage.44,50,51,72 This functional duality is dependent on dmDicer’s helicase domain, but until very recently, we lacked structural insights to explain these activity differences. Using cryo-EM, multiple structures of dmDicer-152 and dmDicer-253,54 in a variety of conformational states have been determined ranging in resolution from 5 to 3 Å. Figure 4 outlines some of these structures in their proposed mechanistic sequence and compares this structural information to what is currently available in regard to hsDicer. Missing from Figure 4 are additional structures of dmDicer-1 in complex with Loqs-PB determined in the presence of Ca2+, as well as dmDicer-1 bound to dsRNA in an intermediate cleavage state,52 a dimer structure of dmDicer-2 in complex with Loqs-PD,54 and apoDmDicer-2 in complex with R2D2.53
Figure 4.
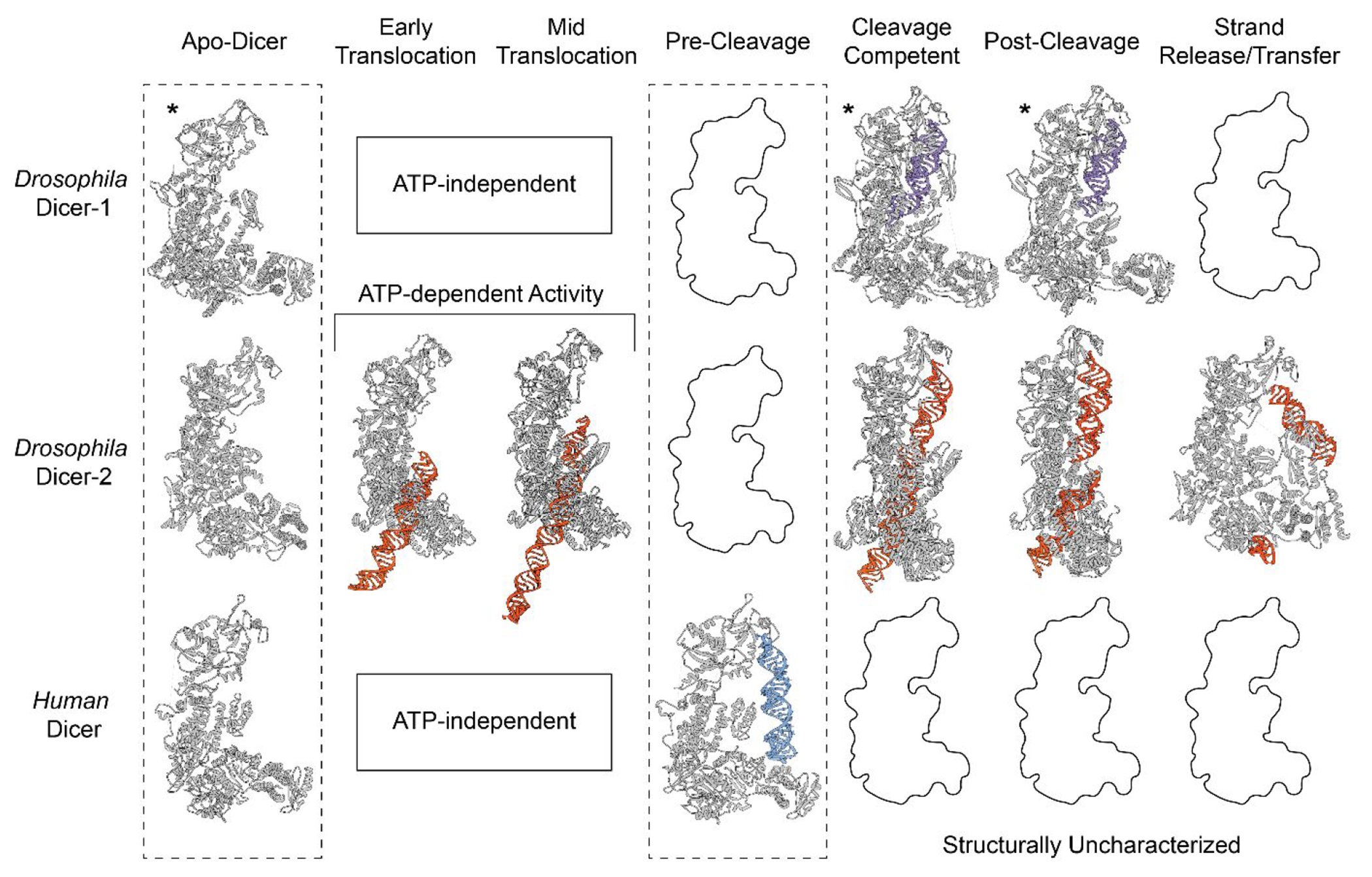
Mechanistic insights into Drosophila and human Dicer through cryo-EM structural studies. Molecular models built from either EM density maps or predicted from AlphaFold* are organized into their proposed mechanistic sequence. The RNA substrates for human Dicer, dmDicer-2, and dmDicer-1 are colored blue, orange, and purple, respectively. Conformations yet to be structurally characterized are left as black outlines. Conformational states that are ATP dependent are only shown for dmDicer-2 (PDBs for Apo-hsDicer: 5ZAK; dmDicer-2: 7W0B; dmDicer-1: 8DG1; Early Translocation dmDicer-2: 7W0C; Midtranslocation dmDicer-2: 7W0D; Pre-Cleavage hsDicer: 5ZAL; Cleavage-Competent dmDicer-2: 7W0E; dmDicer-1: 8DG5; Post-cleavage dmDicer-2: 7W0F; dmDicer-1: 8DGA; Strand Release/Transfer dmDicer-2: 7V6C).
In order to determine the structure of dmDicer-1 in complex with Loqs-PB bound to a pre-miR substrate, a pre-let-7 mimic was used in which the stem region was perfectly base-paired.52 This artificially increased the stability of the RNA substrate in order to capture snapshots of dmDicer-1 in a variety of conformational states. In addition to this pre-let-7 mimic, Ca2+ and Mg2+ ions were used to capture structures of dmDicer-1 in apo- and active states, respectively.52 To enable the active dicing (cleavage-competent) state, an additional step of heating the Dicer-RNA sample for 5 min at 25 °C was performed before vitrification.52 These additional steps, and the use of a highly stable pre-let-7 substrate, allowed for the determination of dmDicer-1 in a cleavage-competent, cleavage intermediate with one RNA strand cleaved and post-cleavage-competent conformations. The apo-, cleavage-competent, and post-cleavage states are shown in Figure 4. Since dmDicer-1 is ATP independent, there are no translocation conformations since that activity is dependent on ATP hydrolysis.
The dmDicer-1 structures present new information regarding the structural basis for dmDicer-1 activity and its interactions with a pre-miR. Based on these structures, the authors suggest that, when not bound to dsRNA, the Platform-PAZ is dynamic which would contribute to the observed lower resolution and loss in density in the EM maps. dmDicer-1 is depicted in cleavage-competent and post-cleavage conformations that seemingly show minimal conformational changes in the helicase domain (Figure 4). These results are surprising when compared to available structures for dmDicer-2 in which there is a large conformational change occurring in the helicase domain moving from an apo- to cleavage-component state (Figure 4). Large conformational changes in the helicase domain have also been reported in cleavage-competent cryo-EM structures of a 3.1 Å DCL3–pre-siRNA complex71 and a recent 6.2 Å mouse Dicer–pre-miR-15a complex.73 Thus, the dmDicer-1 structure suggests that, at least in Drosophila, Dicer may perform its cleavage reactions through an alternative mechanism that is not as strongly dependent on conformational changes in the helicase domain. However, further work is needed using a native pre-miR substrate that has not been modified to determine if these conformational states are biologically relevant.
In contrast to dmDicer-1, which is known to primarily process pre-miRs, dmDicer-2 has an alternative function in processing long dsRNA through the utilization of its ATP-dependent helicase domain.51,74 Structures of dmDicer2 in complex with Loqs-PD bound to a 50 base pair (bp) dsRNA were determined using a catalytically inactive dmDicer-2 mutant (D1217N/D1476N).54 The use of this dmDicer2 mutant, in addition to assembling components in the presence or absence of ATP, resulted in the structural determination of dmDicer-2–Loqs-PD in complex with a 50 bp dsRNA in early translocation, midtranslocation, cleavage-competent, and post-cleavage conformations (Figure 4).54 These structures directly support previous midresolution 7 Å dmDicer-2 structures49 and transient kinetic studies that characterize the importance of ATP for translocation activity.50,51,75–78 Additionally, separate structural studies of dmDicer-2 bound to R2D2 in complex with siRNA revealed structural information for a strand selection/transfer conformation (Figure 4).53 Together, these structures depict a nearly complete stepwise mechanism for dmDicer-2 ATP-dependent activity. However, while these structures may suggest that we have learned all we can about dmDicer-2, a unique dmDicer-2 dimer which is not shown in Figure 4 was also determined, revealing that, in the absence of ATP, dmDicer-2 dimerizes to form an initial binding complex with dsRNA.54 This type of Dicer dimerization has not been previously reported and, even more interestingly, involves an interface at the RNase IIIA insert typically thought to function in Ago2 binding in hsDicer.57 As a result, this structural discovery introduces new questions regarding the biological importance of this dimerization and what additional binding or mechanistic roles the RNase IIIA insert may play.
As more studies continue to advance our structural knowledge of Dicer, it is important to focus on the key questions that remain to be answered. These include filling in the missing structural conformations of hsDicer (Figure 4), uncovering what activates Dicer’s transition from the pre-cleavage to cleavage state, and probing the structural differences of Dicer when bound to a variety of pre-miR and non-pre-miRNA substrates.56,79 As seen in the dmDicer-1 and dmDicer-2 structures, the RNA substrate used can have a major impact on the structural state. As a result, it is important to recognize that current structural studies of hsDicer only depict structures in the presence of a pre-miR substrate. Moving forward, designing structural experiments that contain both pre-miR and nonpre-miR substrates will help us to better understand and model hsDicer’s versatile function(s). Additionally, further characterization of the noncatalytic domains and regions of Dicer is needed to determine if they have additional functions outside of Dicer cleavage activity. In response, the following sections focus on presenting what is known and unknown about hsDicer, as well as how we can utilize currently available Dicer and Dicer-like structures to propose a hypothetical model to explain the hsDicer mechanism-of-action.
OVERVIEW OF HUMAN DICER DOMAINS AND FUNCTIONS
Despite the structural gaps in our knowledge of hsDicer, years of functional studies have given us insight into the individual domains of Dicer and their impact on activity.5,17,80,81 Dicer plays a major role in multiple RNAi pathways, and its sequence is highly conserved across eukaryotes with many species containing more than one copy of Dicer, each with divergent functions.17,80–82 HsDicer contains an N-terminal DRA helicase domain consisting of 3 subdomains: Hel1, Hel2i, and, Hel2. This is followed by a pincer domain, a DUF283 domain, a Platform-PAZ-connector helix (PPC) domain, a large unknown region, two RNase III domains (RNase IIIA and RNase IIIB), and a C-terminal dsRBD (Figure 2). The structural organization of these domains is shown in Figure 5A modeled from the 4.4 Å structure of the apo-Dicer-TRBP complex determined via cryo-EM.70 This model depicts the characteristic L-shaped architecture of hsDicer with the N-terminal helicase domain forming the bottom of the overall L-shape. The pincer domain connects the helicase domain to DUF283, and from there, the linker between DUF283 and the PPC domains, which is missing in this structure, is postulated to connect the N-terminus of the enzyme to the top PPC domain. The PPC domain anchors the top of the enzyme and contains a unique Dicer-specific α-helix that is not found in homologous PAZ domains such as that in Ago2.67,83 Finally, there is an unknown region that connects the PPC from the connector helix to the RNase IIIA and IIIB processing center in the middle of the enzyme and dsRBD at the C-terminus. While the function of this unknown region has yet to be fully determined, recent cryo-EM structures of dmDicer-253,54 have given us some insight into its structural purpose but this region remains unseen in cryo-EM structures of hsDicer.70 The RNase III, PPC, and helicase have been determined to be the primary domains facilitating the binding and cleavage of RNA substrates.34,55,67 In the following section, we discuss what is known about these major functional domains, as well as what remains to be characterized.
Figure 5.
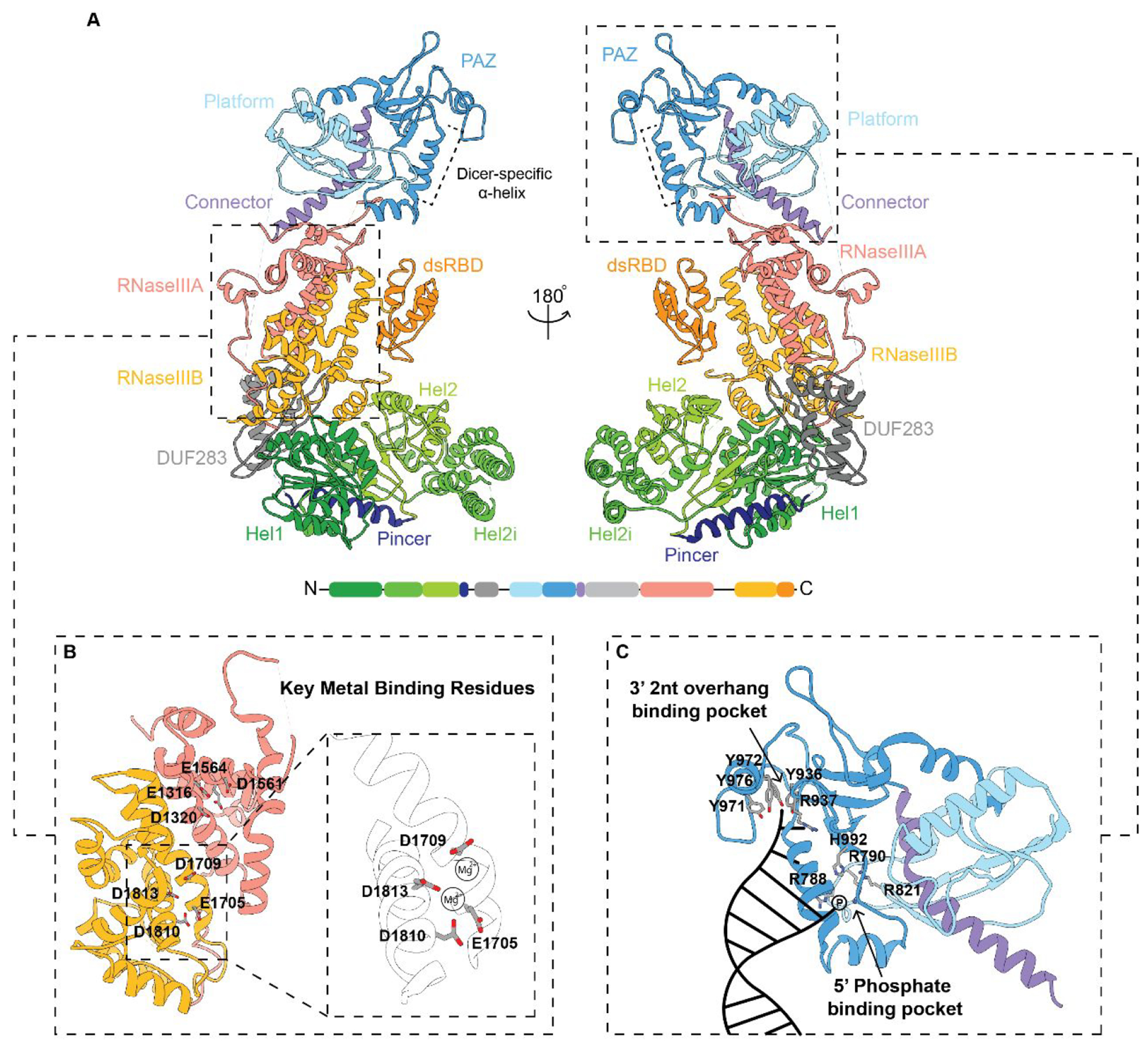
Structural model of human Dicer. (A) Structural overview modeled from a 4.4 Å cryo-EM structure. Domain organization and color code used for labeling the domains from the N- to C-termini are also shown at the bottom. Dashed panels are zoomed-in views of the (B) RNase III processing center and (C) major binding pockets within the Platform-PAZ domains with key amino acids highlighted. A cartoon representation of RNA binding within the 3′ 2 nt overhang and 5′ phosphate binding pockets is depicted in the bottom right panel in (C). (PDB: 5ZAK).
RNase III DOMAINS
The RNase III domain is the defining catalytic domain for all RNase III family enzymes. Mechanistically, each RNase III domain cleaves dsRNA substrates through the coordination of two divalent Mg2+ ions.84 This coordination is used to catalyze the hydrolysis of a phosphodiester bond within each strand of the dsRNA. A network of acidic amino acids found within the RNase III domains are key to stabilizing Mg2+ coordination. In hsDicer, these residues are E1316, D1320, D1561, and E1564 within the RNase IIIA domain and E1705, D1709, D1810, and E1813 within the RNase IIIB domain (Figure 5B).70,81 A proposed model for Mg2+ ion coordination by key metal-binding residues in RNase IIIB is shown in Figure 5B with D1709 and D1813 coordinating one Mg2+ and E1705, D1810, and D1813 binding to the second Mg2+.17,34,71 In the absence of Mg2+, Dicer is unable to perform its cleavage reaction. In fact, studies have shown that the replacement of Mg2+ with Ca2+ can have an inhibitory effect on Dicer cleavage activity.84,85 As such, Ca2+ is often used to study binding affinity in the absence of a chelating agent, such as EDTA, to avoid any unwanted cleavage reaction. Recent structural studies have also utilized the inhibitory effect of replacing Mg2+ for Ca2+ in the RNase III active site as an approach for determining Dicer and Drosha structures bound to their respective uncleaved pre- and pri-miRNA substrates.39,70 However, while this eliminates unwanted substrate cleavage, the effects of ion replacement on RNA substrate stability are not well understood.86,87 Structural methods that focus on profiling inactivating mutations, such as those used to characterized dmDicer-2,54 rather than ion replacement may offer a more accurate depiction of pre-cleavage RNA–protein interactions.
Taking a closer look at the interplay between the RNase IIIA and RNase IIIB domains, biochemical studies have shown that these domains can act independently of each other.88 Class II and Class III enzymes differentiate the cleavage reactions associated with each domain so that the RNase IIIA and RNase IIIB domains cleave the 3P and 5P miRNA strands, respectively.88–90 Mutations that change the metal-binding amino acid residue D1709 in the RNase IIIB domain cause significant negative effects on 5P strand processing leading to a 3P strand bias.88,91 Additionally, mutations at the remaining metal-binding residues within the RNase IIIB domain also lead to a decrease in 5P strand processing but not to the same severity as the D1709 mutation.88 Mutations in the RNase IIIB domain, collectively termed Dicer hotspot mutations (E1705, D1709, D1810, and E1813, Figures 5B and 6),91 are associated with Dicer-related disease onset and an increased risk for developing tumors in the lungs, kidneys, ovaries, thyroid, and several other organs in the human body.92–94 Often referred to as DICER1 Syndrome, this genetic disorder is linked to a loss of 5P strand processing.91,94–96 Disease-associated mutations that cause a loss in the processing of both strands are also common, but mutations that primarily affect 3P strand processing have currently not been linked to the disease.91,97,98 Some examples of disease-associated Dicer mutations that have been shown to affect Dicer processing but are not located in the RNase IIIB domain are S1344L, L881P, and S839F (Figure 6).97–103 These mutations are located within the RNase IIIA domain, the Platform domain, and the PAZ domain, respectively. With respect to the mechanism-of-action of these mutations, the RNase IIIA mutant S1344L is predicted to affect Mg2+ coordination within the RNase IIIB domain and impact 5P strand processing similar to the Dicer hotspot mutations despite its association within the RNase IIIA domain.99 In contrast, the disease-associated mutations L881P and S839F located in the Platform-PAZ domains are located away from both the catalytic site and any binding sites.97,98 As such, these mutations have no direct basis for affecting Dicer processing but have been shown to decrease both 5P and 3P strand processing.97,98 Interesting, a mutation at R944 was also identified in patients but did not result in the development of a disease phenotype despite its close proximity to the RNA binding pockets.99 Further research, both biochemical and structural, is needed to better understand how mutations in domains not directly associated with the RNA processing center of the protein alter Dicer processing of both RNA strands and whether changes in Dicer cleavage activity may vary depending on which RNA substrate is being cleaved.
Figure 6.
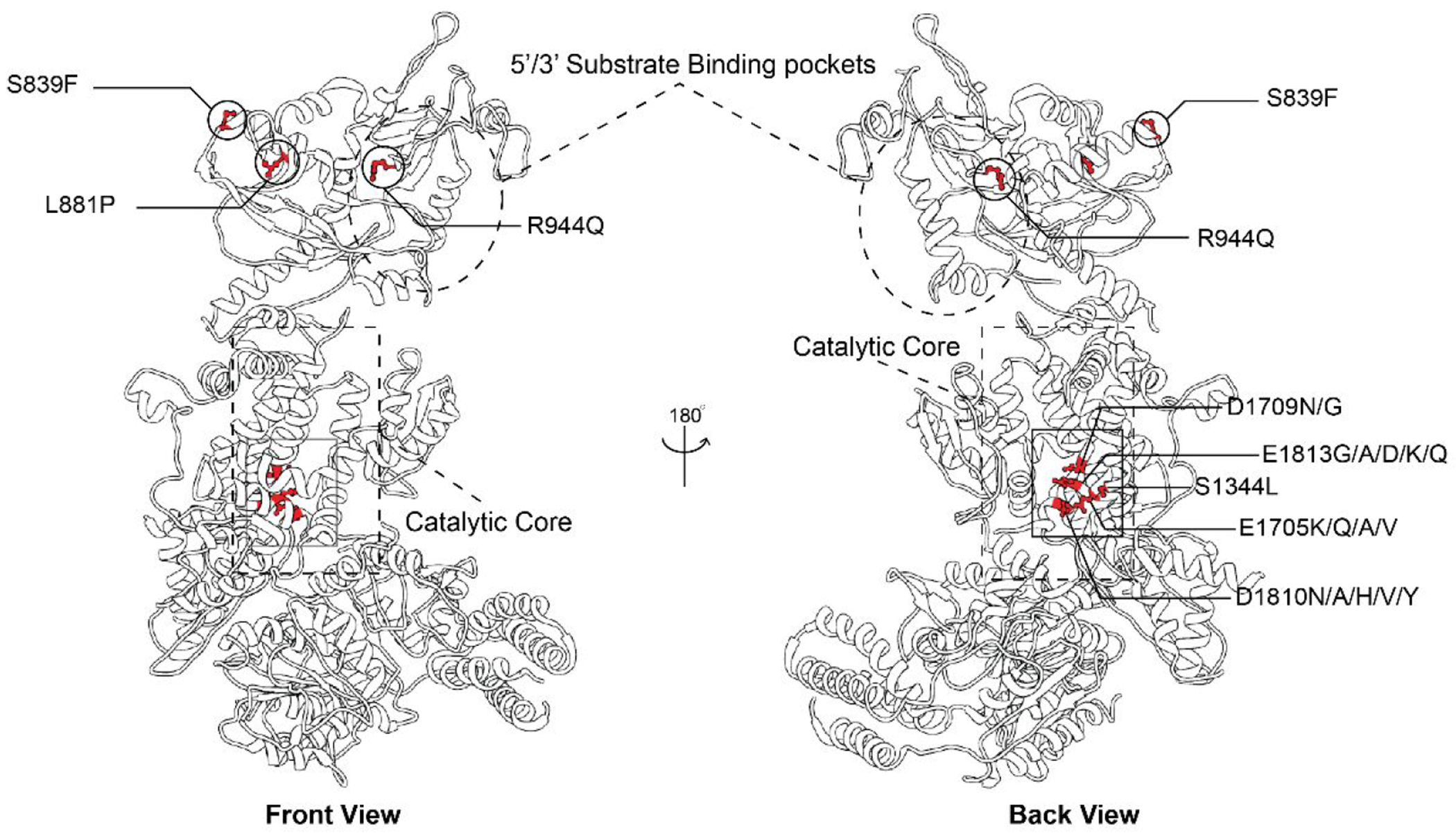
3D structural representation of disease-associated Dicer mutation and their locations relative to major binding and catalytic sites. Key residues are highlighted including metal binding residues (D1709, E1705, D1810, and E1813 within the RNase IIIB domain catalytic core), S1344 located within the RNase IIIA domain catalytic core, and Platform-PAZ domain mutants (S839, L881, and R944) (PDB: 5ZAK).
PLATFORM-PAZ-CONNECTOR DOMAIN
Following Drosha processing, the pre-miRNA substrate contains a 3′ 2nt overhang along with a phosphorylated 5′ end, which are required for recognition by hsDicer, specifically its PAZ and Platform domains, respectively (Figure 5C).42,48,55,67 Within the PAZ domain, the amino acid residues R937, Y936, Y971, Y972, and Y976 form the 3′ 2nt overhang binding pocket. The 5′ phosphate binding pocket, in contrast, is contained primarily within the Platform domain and consists of residues R788, R790, R821, and H992.41,67,70,104 Of note, the residue assignments shown in Figure 5C are based on the sequence reported from the cryo-EM apo-Dicer-TRBP complex structure,70 which is notably 10 amino acids off from the original PPC crystal structure (PDB: 4NGG).67 The importance of these binding pockets has been further explored through biochemical and mutational studies, demonstrating their significance in enabling Dicer’s ability to bind to RNA and initiate the cleavage reaction.67
Compared across species, the Platform-PAZ and RNase III domains are considered the functional core of Dicer-type enzymes since these domains are present in the minimal Dicer found in Giardia and are highly conserved among eukaryotes.26,55 The presence of the Platform-PAZ domain allows Dicer to accurately cleave RNA substrates to a precise, predetermined length. Deletion of this domain leads to indiscriminate cleavage resulting in the production of miRNAs of varying lengths, thereby affecting downstream mRNA targeting.42,105 It is still not fully understood what precise features of Dicer and/or RNA define the substrate length; however, accepted models are that the enzyme uses RNA-binding interactions in the Platform-PAZ and/or helicase domain to determine sites of cleavage (Figure 7).48,55,79
Figure 7.
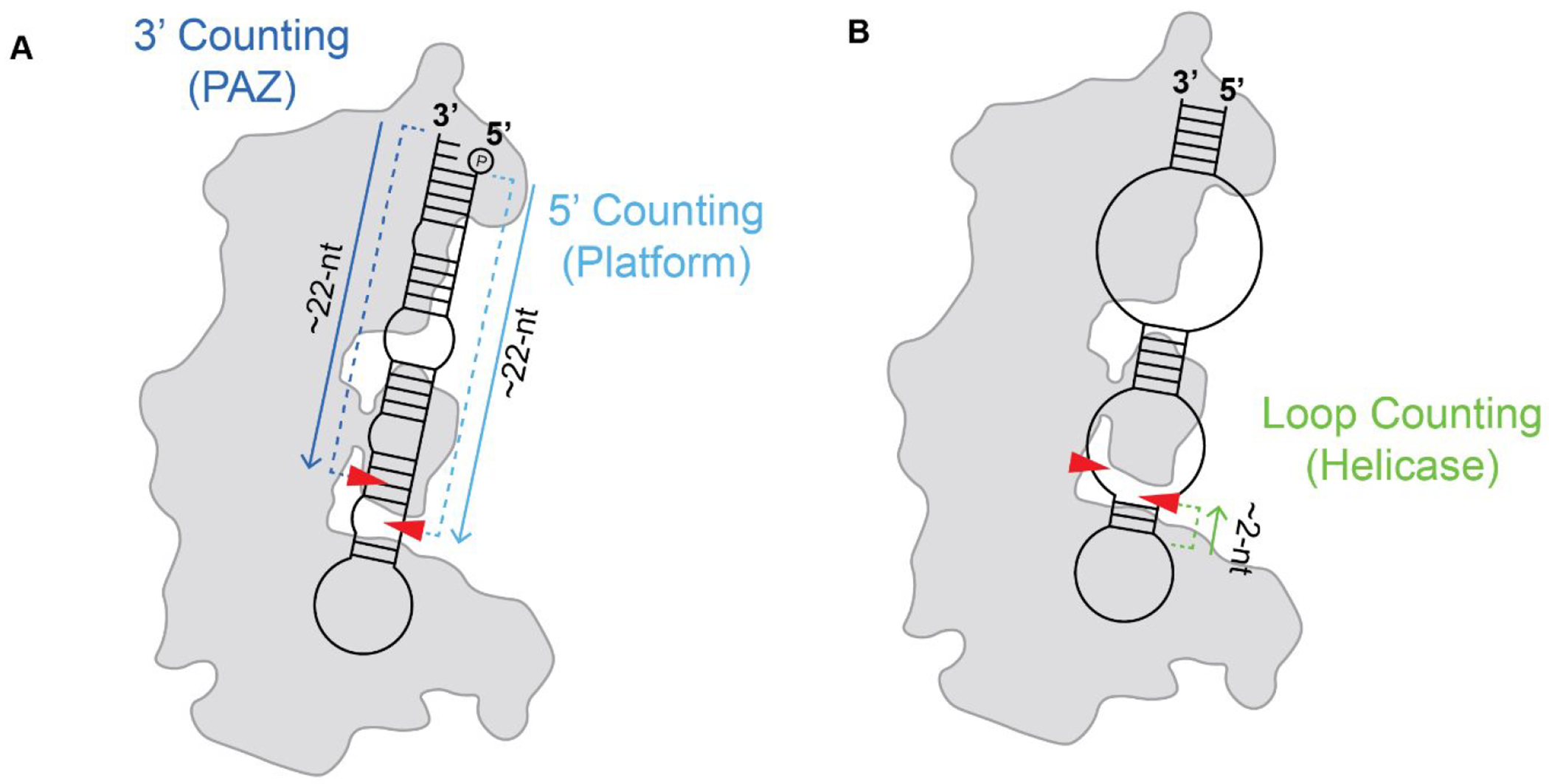
Dicer cleavage counting rules. (A) 5′ and 3′ counting rules determined by RNA binding within the Platform (light blue) and PAZ (dark blue) domains, respectively. (B) Loop counting rule determined by binding within the helicase domain (green). Approximate sites of cleavage are represented by red arrows.
The Dicer counting rule generally refers to the physical distance between RNA binding pockets and the RNase III catalytic residues. These cleavage counting rules are the 3′ counting rule, 5′ counting rule, and loop counting rule (Figure 7).48,49,55,79 In the proposed 3′ counting rule, Dicer measures from the 3′ 2-nt overhang of the RNA substrate toward the loop region cleaving at a length of ~22 nt; while in the 5′ counting rule, the counting to determine the cleavage site starts from the 5′ phosphate (Figure 7A).42,79 In the loop counting rule, Dicer uses the single-stranded regions of the RNA substrate, either an internal bulge or terminal hairpin loop, to anchor the cleavage site 2 nt upstream of this region (Figure 7B).48,79,90 Recent studies using an in vivo click selective 2′-hydroxyl acylation and profiling experiment and mutational profiling (icSHAPE-MaP) surveyed the structural landscape of hsDicer substrates to determine if structural features associated with the RNA substrate determine which counting rule hsDicer employs to measure cleavage product length.79 Interestingly, results from this study revealed that there is likely no one dominant counting rule “enforced” by hsDicer. Instead, in many cases, the factor determining where a substrate is cleaved and which counting rule is used depends upon the structural features of the RNA substrate. Indeed, recent studies have identified pH-dependent impacts on Dicer-mediated processing of select miRNAs with nucleotide base mismatches and base flipping-induced structural arrangements within the RNA stem region leading to different cleavage sites.106–108 These studies also suggest that the cleavage reactions performed by the RNase IIIA and IIIB domains may not occur simultaneously.79,88 As a result, two separate counting rules could be used to determine the cleavage sites for the 5P and 3P strands.79 This cleavage versatility can act as an additional mechanism that Dicer uses to distinguish between RNA substrates, as well as determine strand selection during RISC assembly. Furthermore, the interplay between Dicer binding at the Platform-PAZ and cleavage in the RNase III domains may be more closely linked than previously thought. As previously mentioned, structural studies of dmDicer-152 revealed a high degree of flexibility in the Platform-PAZ domain, but similar conformational flexibility has not been observed for dmDicer-253,54 or hsDicer67,70 structures. However, transient kinetic studies of dmDicer-2 do suggest that the Platform-PAZ domain may play a larger role in the initiation of cleavage and stabilization of RNA substrates.51 To understand the mechanisms for how cleavage sites are regulated and what additional functionals the Platform-PAZ domain might have in hsDicer, additional kinetic and structural investigations that focus on the intermediate steps occurring during hsDicer processing are needed.
Lastly, one interesting feature of the hsDicer PAZ domain that has only begun to be explored is the Dicer-specific α-helix (DSH).67 This helix insertion, unique to the Dicer family, is notably absent from similar PAZ domains such as those found in other RNA processing enzymes like Ago2.27,83 In crystal structures of the Platform-PAZ, the DSH partially occupies the 5′ phosphate binding pocket when the RNA substrate is not fully bound.67 The role of the DSH and why it is unique to Dicer is still being investigated, but structural studies of the Platform-PAZ domain and full-length hsDicer suggest conformational changes within the Platform-PAZ domain are needed in order to free the DSH from the 5′ phosphate binding pocket, thereby allowing the RNA substrate to fully enter and bind. A deeper exploration into this mechanism is described later in this Perspective.
HELICASE DOMAIN
The hsDicer DRA helicase domain also contributes to RNA-binding interaction as well as substrate unwinding45 and consists of 3 subdomains (Hel1, Hel2i, and Hel2) sharing high sequence identity to the super family 2 nucleic acid-dependent ATPase (SF2) helicase family.44–46 Unlike traditional RNA helicases, such as DExD/H-box helicases which are powered through ATP hydrolysis, DRAs rely on conformational changes that occur upon RNA binding to initiate RNA unwinding activity.45,109 This RNA-binding dependence is also reflected in the unique α-helical insertion domain (HEL2i) not found in other DExD/H-box helicases.110–113 However, not all Dicer helicase domains within the Class III RNase III family function independently of ATP. Furthermore, as shown in Figure 2 and as described in previous sections, ATP dependence, conformational flexibility, and substrate specificity within the helicase domain seems to vary among Dicer species. For instance, while cleavage-competent structures of dmDicer-1 depict minimal changes in the helicase domain, cleavage-competent structures of dmDicer-2, DCL3, and mouse Dicer show significant conformational changes in the helicase domain. What is most interesting about these differences is the fact that those structures showing large conformational changes in the helicase domain are not directly related to if the helicase domain is ATP dependent. Both DCL3 and mouse Dicer have ATP-independent helicase domains and yet share more structural similarities to the ATP-dependent dmDicer-2 than the ATP-independent Dicer-1. It remains to be determined if hsDicer shares more similarity to dmDicer-1 or dmDicer-2; however, based on current structural information, it can be inferred that hsDicer may present more similar conformational changes to those seen for DCL3 and mouse Dicer.
Adding even more complexity, hsDicer also functions in the binding and cleavage of a variety of RNA substrates outside of the RNAi pathway.56,79,114–117 Studies have shown that hsDicer and ceDicer are capable of actively cleaving non-pre-miR substrates such as tRNA and small nucleolar (snoRNA), as well as passively binding mRNA and long noncoding (lncRNA) with no cleavage activity.56 Whether the binding and cleavage of these alternative substrates is ATP dependent or if these interactions may induce a different conformational state than the ones previously determined is currently unknown. Structural and functional studies using a variety of small RNA substrates would aid the determination of what RNA-dependent factors may dictate ATP dependence, as well as substrate specific recognition, cleavage, and conformational changes. The diverse functionality of the Dicer helicase domain seems to contribute to the enzyme’s substrate specificity, but there is still much to be learned with respect to the variability of Dicer ATP dependence. Current structures of hsDicer and mouse Dicer lack the resolution needed to accurately characterize and model the helicase domain to develop a molecular model of how substrate selection occurs.118 As a result, models of helicase function rely on structural studies of homologous domains in RIG-1 and MDA5;46,111,112 however, these structures are unable to provide information regarding how conformational changes that may occur in the helicase domain can affect other domains within hsDicer. To answer these questions, mutational studies of the helicase domain within the context of the full-length hsDicer are needed to identify key functional residues. An ideal place to begin these studies would be within the pincer region which is unique to the SF2 helicase family and has been shown to play an important role in conformational movement for the homologous helicase RIG-1.47,48,111,119 An added benefit of these types of mutational studies would be providing additional targets for structural analysis that could aid in “locking” Dicer in different intermediate states that are normally quickly transitioned through during RNA processing. This would ideally help to better resolve missing regions that are not visible in current structures and improve the overall resolution of hsDicer structural intermediates, making it easier to build more precise models.
INFERRING HUMAN DICER MECHANISM, CLEAVAGE ACTIVATION, AND PRODUCT RELEASE
Transitioning from the functional information gained from studying the individual hsDicer domains, it was not until 2018 that the first full-length structures of hsDicer, both unbound and bound to a pre-miRNA substrate, pre-let-7, reached resolutions enabling molecular modeling of the secondary structure.70 From the structure of the apo-hsDicer-TRBP complex, the major hsDicer domains were modeled using previously reported crystal and homology-based models;70 however, there was no density for the RNase IIIA domain insert containing the Ago2 minimal binding motif and the connecting domain between the Platform-PAZ and RNase IIIA. To determine the structure of the hDicer-TRBP-pre-let-7 complex, Ca2+ was used as an ion replacement for Mg2+ to prevent unwanted RNA processing.70,84 Interestingly, the subsequent structure of the hsDicer-pre-miRNA complex reflected a pre-cleavage state with the RNA extended away from the catalytic core (Figure 8). Furthermore, this complex showed the 5′ end of the RNA nearly 15 Å away from its proposed binding pocket, suggesting incomplete binding to the Platform domain (Figure 9C). This finding suggests that an additional step might be needed to activate hsDicer and its transition into a cleavage-competent state. Yet, lack of high-resolution information and missing density in the map for the helicase domain hinders interpretation of how hsDicer binds and interacts with the hairpin loop region of the RNA. While the hsDicer-TRBP complex structures are the first examples of visualizing the 3D organization of full-length hsDicer at resolutions possible to recognize secondary structural information in specific domains, how hsDicer interacts with pre-miRNA in the helicase domain and what additional conformational changes take place to initiate the cleavage reaction remain unknown (Figure 4).
Figure 8.
Structural comparison of cryo-EM densities for the hsDicer pre-cleavage state and DCL3 cleavage-competent state. An overview of domain organization and color code for each protein is shown above. Region/domains missing density are shown in light gray in the domain organization and are represented by dashed lines in the EM structures. For both EM structures, the individual RNA strands are colored gray and dark gray (EMBD: hsDicer-6905; DCL3-31963).
Figure 9.
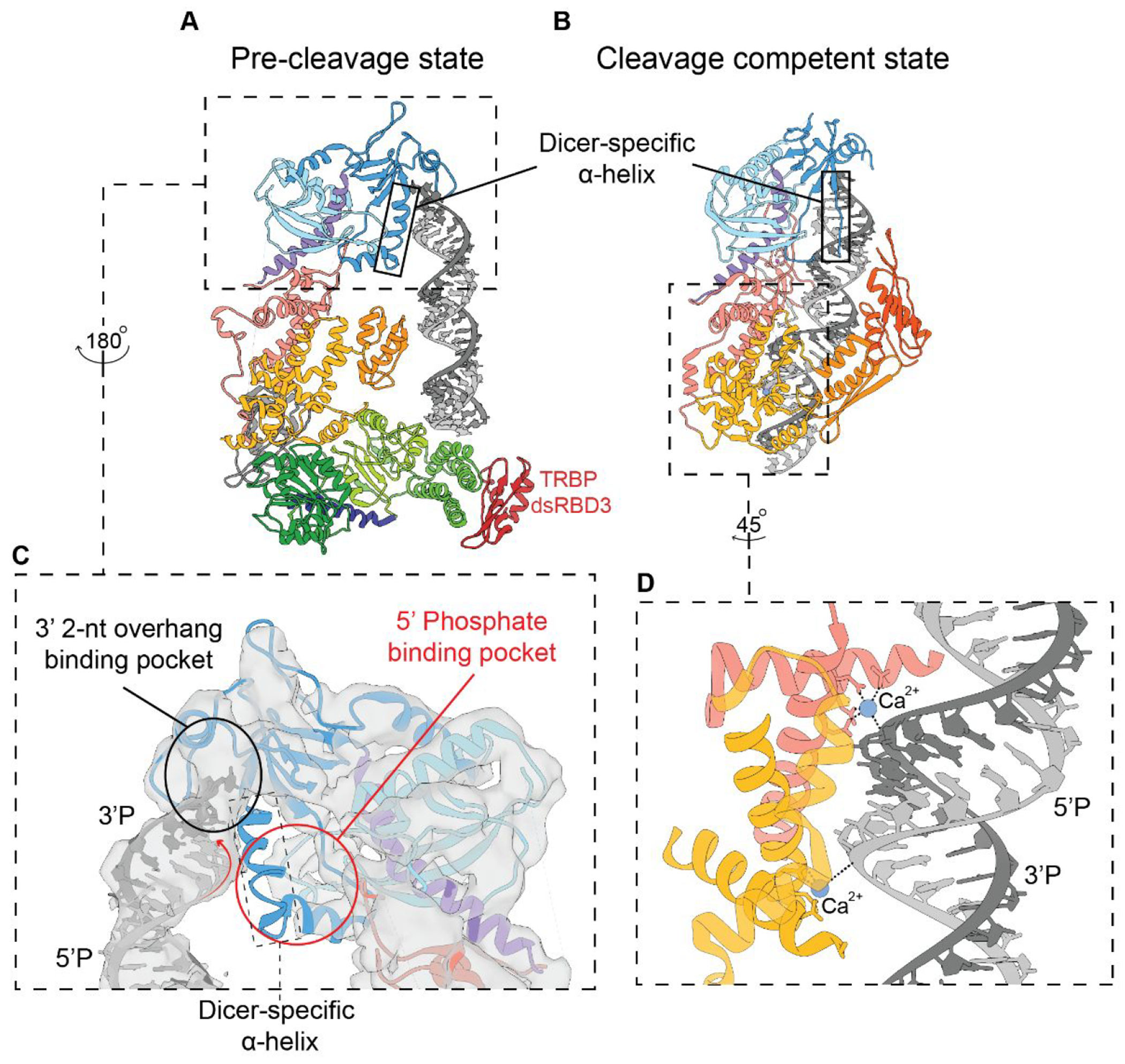
Overview of the major structural differences between the (A) hsDicer pre-cleavage complex and (B) DCL3 cleavage-competent complex cryo-EM structures. Outlined in black is the Dicer-specific α-helix which has a notable structural change from structured to unstructured within the transition between states. (C) Significant binding interactions occurring in the pre-cleavage structure with a red circle indicating the absence of complete binding by the pre-miRNA substrate in the Platform domain. (D) Zoomed in view of the catalytic core of DCL3 with Ca2+ bridging the contact between the RNase III domains and pre-siRNA substrate (PDB: hsDicer, 5ZAL; DCL3, 7VG2).
Following these structural studies, in 2021, a 3.1 Å structure of Arabidopsis (plant) DCL3 was determined using cryo-EM. This study described the first cleavage-competent structure of Dicer bound to a dsRNA substrate at a resolution appropriate for building a detailed molecular model.71 Structures of DCL1 bound to pre-siRNA and pre-miRNA were also determined during this time but at lower resolutions (4.6 and 4.9 Å, respectively).120 The quality of the 3.1 Å DCL3-pre-siRNA complex density map allowed for de novo model building of Dicer and the RNA substrate, revealing for the first time the direct amino acid interactions of Dicer with the RNA substrate.71 In order to prepare a stable DCL3-pre-siRNA complex, Mg2+ was replaced with Ca2+ to mitigate unwanted RNA processing, and an additional cross-linking step was performed before vitrification to promote locking the enzyme in a cleavage-competent conformation.71,121 A similar protocol was followed to determine a 3.7 Å structure of the Drosha-DGCR8 complex bound to a pri-miRNA in a fully docked conformation using cryo-EM.39 Despite its significance, there are still major domains missing from the DCL3 structure (Figure 8). Again, density for the helicase domain was not well resolved in the EM map and is blurred in the 2D class averages. As mentioned previously, a 6.2 Å cryo-EM structure of mouse Dicer bound to pre-miR-15a in a cleavage-competent conformation also showed a loss in density within the helicase domain and has a similar overall structure compared to DCL3.73,109 This lack of density for the helicase domain in multiple cryo-EM structures from multiple organisms suggests extensive flexibility in this domain and/or that a large conformational change occurs in this region during the cleavage process. More work remains to find ways to stabilize the helicase domain in the cleavage-competent state.
By comparing the structures of hsDicer and DCL3, we can start to propose a mechanism for understanding and explaining hsDicer function. We focus on DLC3 rather than the other structures of Dicer due to its overall structurally similarity to the cleavage-competent mouse Dicer cryo-EM structure73 and since DLC3 contains an ATP-independent helicase.71 Furthermore, while the mouse Dicer shares far more similarities in size and sequence to hsDicer,32,122 at 3.1 Å, the DCL3-siRNA cleavage-competent structure gives us much more structural information to learn from compared to the 6.2 Å mouse Dicer structure. Lastly, it should be noted that both the DCL3 and mouse Dicer structures are lacking EM density for the helicase domain in their cleavage-competent conformations, making it difficult to infer the mechanism for cleavage activation from this structural information alone. Therefore, we focused these comparisons around subtle changes in Platform-PAZ domain, specifically in reference to the Dicer-specific α-helix (DSH). As shown in Figure 9, in transitioning from the pre-cleavage to cleavage-competent state, the DSH loses its overall α-helical structure and becomes disordered. In previous X-ray crystallographic studies of the Platform-PAZ bound to siRNA substrates, the DSH was found to separate the two RNA-binding pockets and orient the bound siRNA away from Dicer.67 A subset of the structures revealed a second conformation in which this DSH was disordered with the bound siRNA aligned toward Dicer (PDB: 4NHA).67 These studies categorized the ordered and disordered DSH structures as product release/transfer and cleavage-competent complexes, respectively.
Since these X-ray crystallographic studies focused solely on a single domain (e.g., PPC), it is difficult to determine the purpose of this order-to-disorder transition; however, now observing a similar structural change within the context of full-length Dicer and Dicer-like structures, a model explaining the importance of this ordered-to-disordered change can be developed. In both cleavage-competent structures, the DSH is disordered with the RNA substrate ideally positioned for cleavage. When the DSH becomes ordered, as shown in the pre-cleavage hsDicer and proposed Platform-PAZ release/transfer structures,67 the DSH occupies the space between the 3′ and 5′ RNA binding pockets inhibiting binding of the 5′ phosphate. Using these structures as a guide, we propose that the transition from pre-cleavage to cleavage-competent and product release/transfer states is dependent upon the structural order and position of the DSH. This proposed transition state mechanism suggests a release of the 5′ phosphate associated with the 5P strand but retention of the 3′ overhang of the 3P strand in the pre-cleavage state in which the DSH is still structured. Upon binding within the 5′ phosphate pocket, the DSH denatures allowing for complete phosphate binding and transition to a cleavage-competent state. After the cleavage reaction and once the product is ready for release, the DSH reoccupies the 5′ pocket and kicks out the 5′ phosphate group of the 5P strand. Overall, this comparison suggests that the DSH plays a role in priming and releasing RNA substrates; however, further structural studies to isolate a cleavage-competent conformation of hsDicer and functional studies determining how the DSH might affect product release are needed to better model this proposed mechanism.
CONCLUSIONS
While the structures described above have provided essential molecular models that make it possible for the first time to propose detailed models for dmDicer-1 and dmDicer-2 functions, key questions still remain regarding hsDicer. These include how the various domains function in the context of the holoenzyme before, during, and after the cleavage reaction. Most notably, current structures of hsDicer have yet to be able to resolve the helicase domain or the various conformational changes likely required for initiating binding and cleavage of RNA substrates. To date, there is only one partial crystal structure of HEL2i bound to one of the dsRBD of TRBP, and much of what we know about the human helicase domain is due to extrapolating from homologous protein structures.47,111,113,118 Furthermore, structures of the RNase III domains have focused primarily on studying a homodimer between RNase IIIB domains, giving us limited understanding of the RNase IIIA domain and its Ago2 minimal binding motif. Answering these questions and aiming to resolve intermediate states of hsDicer bound to a variety of RNA substrates are necessary to generate a detailed model of hsDicer function, including how the enzyme chooses and processes various RNA substrates. Until then, we will continue to struggle in defining hsDicer RNA specificity and ways in which this process can be controlled. However, once we have made such breakthroughs, a new and exciting route toward the development of miRNA-targeted therapeutics will be introduced. From there, structure-guided drug discovery can be deployed to target critical regulatory regions of the Dicer family to therapeutically manipulate gene silencing mechanisms for the treatment of diseases.
Funding
This work was supported by the National Institutes of Health (R01 GM135252 to A.L.G. and F31 GM139291 to R.M.T.).
Footnotes
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.biochem.2c00570
The authors declare no competing financial interest.
Contributor Information
Rachel M. Torrez, Department of Medicinal Chemistry, College of Pharmacy, University of Michigan, Ann Arbor, Michigan 48109, United States; Life Sciences Institute, University of Michigan, Ann Arbor, Michigan 48109, United States
Melanie D. Ohi, Life Sciences Institute, University of Michigan, Ann Arbor, Michigan 48109, United States; Department of Cell and Developmental Biology, University of Michigan Medical School, Ann Arbor, Michigan 48109, United States.
Amanda L. Garner, Department of Medicinal Chemistry, College of Pharmacy, University of Michigan, Ann Arbor, Michigan 48109, United States.
REFERENCES
- (1).Treiber T; Treiber N; Meister G Regulation of MicroRNA Biogenesis and Its Crosstalk with Other Cellular Pathways. Nat. Rev. Mol. Cell Biol 2019, 20 (1), 5–20. [DOI] [PubMed] [Google Scholar]
- (2).Friedman RC; Farh KK-H; Burge CB; Bartel DP Most Mammalian MRNAs Are Conserved Targets of MicroRNAs. Genome Res. 2009, 19 (1), 92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Bartel DP Metazoan MicroRNAs. Cell 2018, 173 (1), 20–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Ha M; Kim VN Regulation of MicroRNA Biogenesis. Nat. Rev. Mol. Cell Biol 2014, 15 (8), 509–524. [DOI] [PubMed] [Google Scholar]
- (5).Yoshida T; Asano Y; Ui-Tei K Modulation of MicroRNA Processing by Dicer via Its Associated DsRNA Binding Proteins. Noncoding RNA 2021, 7 (3), 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Bartel DP MicroRNAs: Target Recognition and Regulatory Functions. Cell 2009, 136 (2), 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Svoboda P Key Mechanistic Principles and Considerations Concerning RNA Interference. Frontiers in Plant Science 2020, 11, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Stavast CJ; Erkeland SJ The Non-Canonical Aspects of MicroRNAs: Many Roads to Gene Regulation. Cells 2019, 8 (11), 1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Yang J-S; Lai EC Alternative MiRNA Biogenesis Pathways and the Interpretation of Core MiRNA Pathway Mutants. Mol. Cell 2011, 43 (6), 892–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).O'Brien J; Hayder H; Zayed Y; Peng C Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Frontiers in Endocrinology 2018, 9, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Peng Y; Croce CM The Role of MicroRNAs in Human Cancer. Sig Transduct Target Ther 2016, 1 (1), 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Rupaimoole R; Slack FJ MicroRNA Therapeutics: Towards a New Era for the Management of Cancer and Other Diseases. Nat. Rev. Drug Discov 2017, 16 (3), 203–222. [DOI] [PubMed] [Google Scholar]
- (13).Li Z; Rana TM Therapeutic Targeting of MicroRNAs: Current Status and Future Challenges. Nat. Rev. Drug Discov 2014, 13 (8), 622–638. [DOI] [PubMed] [Google Scholar]
- (14).Lorenz DA; Garner AL Approaches for the Discovery of Small Molecule Ligands Targeting MicroRNAs. In RNA Therapeutics; Garner AL, Ed.; Topics in Medicinal Chemistry; Springer International Publishing: Cham, 2018; pp 79–110; DOI: 10.1007/7355_2017_3. [DOI] [Google Scholar]
- (15).Disney MD; Suresh BM; Benhamou RI; Childs-Disney JL Progress toward the Development of the Small Molecule Equivalent of Small Interfering RNA. Curr. Opin. Chem. Biol 2020, 56, 63–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Angelbello AJ; Chen JL; Childs-Disney JL; Zhang P; Wang Z-F; Disney MD Using Genome Sequence to Enable the Design of Medicines and Chemical Probes. Chem. Rev 2018, 118 (4), 1599–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Paturi S; Deshmukh MV A Glimpse of “Dicer Biology” Through the Structural and Functional Perspective. Frontiers in Molecular Biosciences 2021, 8, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Leitão AL; Enguita FJ A Structural View of MiRNA Biogenesis and Function. Noncoding RNA 2022, 8 (1), 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Kim VN MicroRNA Biogenesis: Coordinated Cropping and Dicing. Nat. Rev. Mol. Cell Biol 2005, 6 (5), 376–385. [DOI] [PubMed] [Google Scholar]
- (20).Medley JC; Panzade G; Zinovyeva AY MicroRNA Strand Selection: Unwinding the Rules. Wiley Interdiscip Rev. RNA 2021, 12 (3), e1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Chipman LB; Pasquinelli AE MiRNA Targeting: Growing beyond the Seed. Trends in Genetics 2019, 35 (3), 215–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Ui-Tei K; Nishi K; Takahashi T; Nagasawa T Thermodynamic Control of Small RNA-Mediated Gene Silencing. Frontiers in Genetics 2012, 3, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Hibio N; Hino K; Shimizu E; Nagata Y; Ui-Tei K Stability of MiRNA 5′terminal and Seed Regions Is Correlated with Experimentally Observed MiRNA-Mediated Silencing Efficacy. Sci. Rep 2012, 2 (1), 996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Noland CL; Ma E; Doudna JA SiRNA Repositioning for Guide Strand Selection by Human Dicer Complexes. Mol. Cell 2011, 43 (1), 110–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Suzuki HI; Katsura A; Yasuda T; Ueno T; Mano H; Sugimoto K; Miyazono K Small-RNA Asymmetry Is Directly Driven by Mammalian Argonautes. Nat. Struct Mol. Biol 2015, 22 (7), 512–521. [DOI] [PubMed] [Google Scholar]
- (26).Gao Z; Wang M; Blair D; Zheng Y; Dou Y Phylogenetic Analysis of the Endoribonuclease Dicer Family. PLoS One 2014, 9 (4), e95350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Müller M; Fazi F; Ciaudo C Argonaute Proteins: From Structure to Function in Development and Pathological Cell Fate Determination. Frontiers in Cell and Developmental Biology 2020, 7, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Schirle NT; Sheu-Gruttadauria J; MacRae IJ Structural Basis for MicroRNA Targeting. Science 2014, 346 (6209), 608–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Chandradoss SD; Schirle NT; Szczepaniak M; MacRae IJ; Joo C A Dynamic Search Process Underlies MicroRNA Targeting. Cell 2015, 162 (1), 96–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).MacRae IJ; Doudna JA Ribonuclease Revisited: Structural Insights into Ribonuclease III Family Enzymes. Curr. Opin Struct Biol 2007, 17 (1), 138–145. [DOI] [PubMed] [Google Scholar]
- (31).Ciechanowska K; Pokornowska M; Kurzyńska-Kokorniak A Genetic Insight into the Domain Structure and Functions of Dicer-Type Ribonucleases. IJMS 2021, 22 (2), 616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Mukherjee K; Campos H; Kolaczkowski B Evolution of Animal and Plant Dicers: Early Parallel Duplications and Recurrent Adaptation of Antiviral RNA Binding in Plants. Mol. Biol. Evol 2013, 30 (3), 627–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Drider D; Condon C The Continuing Story of Endoribonuclease III. MIP 2005, 8 (4), 195–200. [DOI] [PubMed] [Google Scholar]
- (34).Takeshita D; Zenno S; Lee WC; Nagata K; Saigo K; Tanokura M Homodimeric Structure and Double-Stranded RNA Cleavage Activity of the C-Terminal RNase III Domain of Human Dicer. J. Mol. Biol 2007, 374 (1), 106–120. [DOI] [PubMed] [Google Scholar]
- (35).Zhang H; Kolb FA; Jaskiewicz L; Westhof E; Filipowicz W Single Processing Center Models for Human Dicer and Bacterial RNase III. Cell 2004, 118 (1), 57–68. [DOI] [PubMed] [Google Scholar]
- (36).Gan J; Tropea JE; Austin BP; Court DL; Waugh DS; Ji X Structural Insight into the Mechanism of Double-Stranded RNA Processing by Ribonuclease III. Cell 2006, 124 (2), 355–366. [DOI] [PubMed] [Google Scholar]
- (37).Blaszczyk J; Tropea JE; Bubunenko M; Routzahn KM; Waugh DS; Court DL; Ji X Crystallographic and Modeling Studies of RNase III Suggest a Mechanism for Double-Stranded RNA Cleavage. Structure 2001, 9 (12), 1225–1236. [DOI] [PubMed] [Google Scholar]
- (38).Kwon SC; Nguyen TA; Choi Y-G; Jo MH; Hohng S; Kim VN; Woo J-S Structure of Human DROSHA. Cell 2016, 164 (1), 81–90. [DOI] [PubMed] [Google Scholar]
- (39).Partin AC; Zhang K; Jeong B-C; Herrell E; Li S; Chiu W; Nam Y Cryo-EM Structures of Human Drosha and DGCR8 in Complex with Primary MicroRNA. Mol. Cell 2020, 78 (3), 411–422.E4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Aharoni R; Tobi D Dynamical Comparison between Drosha and Dicer Reveals Functional Motion Similarities and Dissimilarities. PLoS One 2019, 14 (12), e0226147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Ma J-B; Ye K; Patel DJ Structural Basis for Overhang-Specific Small Interfering RNA Recognition by the PAZ Domain. Nature 2004, 429 (6989), 318–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Park J-E; Heo I; Tian Y; Simanshu DK; Chang H; Jee D; Patel DJ; Kim VN Dicer Recognizes the 5′ End of RNA for Efficient and Accurate Processing. Nature 2011, 475 (7355), 201–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Kidwell MA; Chan JM; Doudna JA Evolutionarily Conserved Roles of the Dicer Helicase Domain in Regulating RNA Interference Processing. J. Biol. Chem 2014, 289 (41), 28352–28362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).HANSEN SR; ADEROUNMU AM; DONELICK HM; BASS BL Dicer’s Helicase Domain: A Meeting Place for Regulatory Proteins. Cold Spring Harb Symp. Quant Biol 2019, 84, 185–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Luo D; Kohlway A; Pyle AM Duplex RNA Activated ATPases (DRAs). RNA Biol. 2013, 10 (1), 111–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Zou J; Chang M; Nie P; Secombes CJ Origin and Evolution of the RIG-I like RNA Helicase Gene Family. BMC Evol Biol. 2009, 9, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Ma E; MacRae IJ; Kirsch JF; Doudna JA Auto-Inhibition of Human Dicer by Its Internal Helicase Domain. J. Mol. Biol 2008, 380 (1), 237–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Gu S; Jin L; Zhang Y; Huang Y; Zhang F; Valdmanis PN; Kay MA The Loop Position of ShRNAs and Pre-MiRNAs Is Critical for the Accuracy of Dicer Processing in Vivo. Cell 2012, 151 (4), 900–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Sinha NK; Iwasa J; Shen PS; Bass BL Dicer Uses Distinct Modules for Recognizing DsRNA Termini. Science 2018, 359 (6373), 329–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Welker NC; Maity TS; Ye X; Aruscavage PJ; Krauchuk AA; Liu Q; Bass BL Dicer’s Helicase Domain Discriminates DsRNA Termini to Promote an Altered Reaction Mode. Mol. Cell 2011, 41 (5), 589–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Singh RK; Jonely M; Leslie E; Rejali NA; Noriega R; Bass BL Transient Kinetic Studies of the Antiviral Drosophila Dicer-2 Reveal Roles of ATP in Self–Nonself Discrimination. eLife 2021, 10, e65810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Jouravleva K; Golovenko D; Demo G; Dutcher RC; Hall TMT; Zamore PD; Korostelev AA Structural Basis of MicroRNA Biogenesis by Dicer-1 and Its Partner Protein Loqs-PB. Molecular Cell 2022, 82 (21), 4049–4063.E6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Yamaguchi S; Naganuma M; Nishizawa T; Kusakizako T; Tomari Y; Nishimasu H; Nureki O Structure of the Dicer-2–R2D2 Heterodimer Bound to a Small RNA Duplex. Nature 2022, 607 (7918), 393–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Su S; Wang J; Deng T; Yuan X; He J; Liu N; Li X; Huang Y; Wang H-W; Ma J Structural Insights into DsRNA Processing by Drosophila Dicer-2–Loqs-PD. Nature 2022, 607 (7918), 399–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).MacRae IJ; Zhou K; Li F; Repic A; Brooks AN; Cande WZ; Adams PD; Doudna JA Structural Basis for Double-Stranded RNA Processing by Dicer. Science 2006, 311 (5758), 195–198. [DOI] [PubMed] [Google Scholar]
- (56).Rybak-Wolf A; Jens M; Murakawa Y; Herzog M; Landthaler M; Rajewsky N A Variety of Dicer Substrates in Human and C. Elegans. Cell 2014, 159 (5), 1153–1167. [DOI] [PubMed] [Google Scholar]
- (57).Sasaki T; Shimizu N Evolutionary Conservation of a Unique Amino Acid Sequence in Human DICER Protein Essential for Binding to Argonaute Family Proteins. Gene 2007, 396 (2), 312–320. [DOI] [PubMed] [Google Scholar]
- (58).Tahbaz N; Kolb FA; Zhang H; Jaronczyk K; Filipowicz W; Hobman TC Characterization of the Interactions between Mammalian PAZ PIWI Domain Proteins and Dicer. EMBO Rep 2004, 5 (2), 189–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Chendrimada TP; Gregory RI; Kumaraswamy E; Norman J; Cooch N; Nishikura K; Shiekhattar R TRBP Recruits the Dicer Complex to Ago2 for MicroRNA Processing and Gene Silencing. Nature 2005, 436 (7051), 740–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Kühlbrandt W The Resolution Revolution. Science 2014, 343 (6178), 1443–1444. [DOI] [PubMed] [Google Scholar]
- (61).Frank J Advances in the Field of Single-Particle Cryo-Electron Microscopy over the Last Decade. Nat. Protoc 2017, 12 (2), 209–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Takizawa Y; Binshtein E; Erwin AL; Pyburn TM; Mittendorf KF; Ohi MD While the Revolution Will Not Be Crystallized, Biochemistry Reigns Supreme. Protein Sci. 2017, 26 (1), 69–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Bernstein E; Caudy AA; Hammond SM; Hannon GJ Role for a Bidentate Ribonuclease in the Initiation Step of RNA Interference. Nature 2001, 409 (6818), 363–366. [DOI] [PubMed] [Google Scholar]
- (64).Lyumkis D Challenges and Opportunities in Cryo-EM Single-Particle Analysis. J. Biol. Chem 2019, 294 (13), 5181–5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Peplow M Cryo-Electron Microscopy Reaches Resolution Milestone. ACS Cent. Sci 2020, 6 (8), 1274–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Wu H; Henras A; Chanfreau G; Feigon J Structural Basis for Recognition of the AGNN Tetraloop RNA Fold by the Double-Stranded RNA-Binding Domain of Rnt1p RNase III. Proc. Natl. Acad. Sci. U. S. A 2004, 101 (22), 8307–8312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Tian Y; Simanshu DK; Ma J-B; Park J-E; Heo I; Kim VN; Patel DJ A Phosphate-Binding Pocket within the Platform-PAZ-Connector Helix Cassette of Human Dicer. Mol. Cell 2014, 53 (4), 606–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Lau P-W; Guiley KZ; De N; Potter CS; Carragher B; MacRae IJ The Molecular Architecture of Human Dicer. Nat. Struct Mol. Biol 2012, 19 (4), 436–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Lau P-W; Potter CS; Carragher B; MacRae IJ Structure of the Human Dicer-TRBP Complex by Electron Microscopy. Structure 2009, 17 (10), 1326–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Liu Z; Wang J; Cheng H; Ke X; Sun L; Zhang QC; Wang H-W Cryo-EM Structure of Human Dicer and Its Complexes with a Pre-MiRNA Substrate. Cell 2018, 173 (5), 1191–1203.E12. [DOI] [PubMed] [Google Scholar]
- (71).Wang Q; Xue Y; Zhang L; Zhong Z; Feng S; Wang C; Xiao L; Yang Z; Harris CJ; Wu Z; Zhai J; Yang M; Li S; Jacobsen SE; Du J Mechanism of SiRNA Production by a Plant Dicer-RNA Complex in Dicing-Competent Conformation. Science 2021, 374 (6571), 1152–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Sinha NK; Trettin KD; Aruscavage PJ; Bass BL Drosophila Dicer-2 Cleavage Is Mediated by Helicase- and DsRNA Termini-Dependent States That Are Modulated by Loquacious-PD. Mol. Cell 2015, 58 (3), 406–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Zapletal D; Taborska E; Pasulka J; Malik R; Kubicek K; Zanova M; Much C; Sebesta M; Buccheri V; Horvat F; Jenickova I; Prochazkova M; Prochazka J; Pinkas M; Novacek J; Joseph DF; Sedlacek R; Bernecky C; O’Carroll D; Stefl R; Svoboda P Structural and Functional Basis of Mammalian MicroRNA Biogenesis by Dicer. Mol. Cell 2022, 82 (21), 4064–4079.E13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Lee YS; Nakahara K; Pham JW; Kim K; He Z; Sontheimer EJ; Carthew RW Distinct Roles for Drosophila Dicer-1 and Dicer-2 in the SiRNA/MiRNA Silencing Pathways. Cell 2004, 117 (1), 69–81. [DOI] [PubMed] [Google Scholar]
- (75).Jonely M; Singh RK; Donelick HM; Bass BL; Noriega R Loquacious-PD Regulates the Terminus-Dependent Molecular Recognition of Dicer-2 toward Double-Stranded RNA. Chem. Commun 2021, 57 (83), 10879–10882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Trettin KD; Sinha NK; Eckert DM; Apple SE; Bass BL Loquacious-PD Facilitates Drosophila Dicer-2 Cleavage through Interactions with the Helicase Domain and DsRNA. Proc. Natl. Acad. Sci. U. S. A 2017, 114 (38), E7939–E7948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Donelick HM; Talide L; Bellet M; Aruscavage PJ; Lauret E; Aguiar ERGR; Marques JT; Meignin C; Bass BL In Vitro Studies Provide Insight into Effects of Dicer-2 Helicase Mutations in Drosophila Melanogaster. RNA 2020, 26 (12), 1847–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Hansen SR; Aderounmu AM; Donelick HM; Bass BL Dicer’s Helicase Domain: A Meeting Place for Regulatory Proteins. Cold Spring Harb Symp. Quant Biol 2019, 84, 185–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Luo Q-J; Zhang J; Li P; Wang Q; Zhang Y; Roy-Chaudhuri B; Xu J; Kay MA; Zhang QC RNA Structure Probing Reveals the Structural Basis of Dicer Binding and Cleavage. Nat. Commun 2021, 12 (1), 3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Kurzynska-Kokorniak A; Koralewska N; Pokornowska M; Urbanowicz A; Tworak A; Mickiewicz A; Figlerowicz M The Many Faces of Dicer: The Complexity of the Mechanisms Regulating Dicer Gene Expression and Enzyme Activities. Nucleic Acids Res. 2015, 43 (9), 4365–4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Vergani-Junior CA; Tonon-da-Silva G; Inan MD; Mori MA DICER: Structure, Function, and Regulation. Biophys Rev. 2021, 13 (6), 1081–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Ketting RF; Fischer SEJ; Bernstein E; Sijen T; Hannon GJ; Plasterk RHA Dicer Functions in RNA Interference and in Synthesis of Small RNA Involved in Developmental Timing in C. Elegans. Genes Dev 2001, 15 (20), 2654–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Faehnle CR; Elkayam E; Haase AD; Hannon GJ; Joshua-Tor L The Making of a Slicer: Activation of Human Argonaute-1. Cell Reports 2013, 3 (6), 1901–1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Sun W; Pertzev A; Nicholson AW Catalytic Mechanism of Escherichia Coli Ribonuclease III: Kinetic and Inhibitor Evidence for the Involvement of Two Magnesium Ions in RNA Phosphodiester Hydrolysis. Nucleic Acids Res. 2005, 33 (3), 807–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Li HL; Chelladurai BS; Zhang K; Nicholson AW Ribonuclease III Cleavage of a Bacteriophage T7 Processing Signal. Divalent Cation Specificity, and Specific Anion Effects. Nucleic Acids Res. 1993, 21 (8), 1919–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Misra VK; Draper DE On the Role of Magnesium Ions in RNA Stability. Biopolymers 1998, 48 (2–3), 113–135. [DOI] [PubMed] [Google Scholar]
- (87).DRAPER DE A Guide to Ions and RNA Structure. RNA 2004, 10 (3), 335–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (88).Gurtan AM; Lu V; Bhutkar A; Sharp PA In Vivo Structure–Function Analysis of Human Dicer Reveals Directional Processing of Precursor MiRNAs. RNA 2012, 18 (6), 1116–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (89).Ando Y; Maida Y; Morinaga A; Burroughs AM; Kimura R; Chiba J; Suzuki H; Masutomi K; Hayashizaki Y Two-Step Cleavage of Hairpin RNA with 5’ Overhangs by Human DICER. BMC Molecular Biology 2011, 12 (1), 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (90).Feng Y; Zhang X; Graves P; Zeng Y A Comprehensive Analysis of Precursor MicroRNA Cleavage by Human Dicer. RNA 2012, 18 (11), 2083–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (91).Anglesio M; Wang Y; Yang W; Senz J; Wan A; Heravi-Moussavi A; Salamanca C; Maines-Bandiera S; Huntsman D; Morin G Cancer-Associated Somatic DICER1 Hotspot Mutations Cause Defective MiRNA Processing and Reverse-Strand Expression Bias to Predominantly Mature 3p Strands through Loss of 5p Strand Cleavage. Journal of Pathology 2013, 229 (3), 400–409. [DOI] [PubMed] [Google Scholar]
- (92).Kim J; Schultz KAP; Hill DA; Stewart DR The Prevalence of Germline DICER1 Pathogenic Variation in Cancer Populations. Mol. Genet Genomic Med 2019, 7 (3), e555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (93).Kim J; Field A; Schultz KAP; Hill DA; Stewart DR The Prevalence of DICER1 Pathogenic Variation in Population Databases. Int. J. Cancer 2017, 141 (10), 2030–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (94).Robertson JC; Jorcyk CL; Oxford JT DICER1 Syndrome: DICER1Mutations in Rare Cancers. Cancers (Basel) 2018, 10 (5), 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (95).Foulkes WD; Priest JR; Duchaine TF DICER1: Mutations, MicroRNAs and Mechanisms. Nat. Rev. Cancer 2014, 14 (10), 662–672. [DOI] [PubMed] [Google Scholar]
- (96).Theotoki EI; Pantazopoulou VI; Georgiou S; Kakoulidis P ; Filippa V; Stravopodis DJ; Anastasiadou E Dicing the Disease with Dicer: The Implications of Dicer Ribonuclease in Human Pathologies. International Journal of Molecular Sciences 2020, 21 (19), 7223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (97).de Kock L; Wu MK; Foulkes WD Ten Years of DICER1Mutations: Provenance, Distribution, and Associated Phe-notypes. Human Mutation 2019, 40 (11), 1939–1953. [DOI] [PubMed] [Google Scholar]
- (98).Frio TR; Bahubeshi A; Kanellopoulou C; Hamel N; Niedziela M; Sabbaghian N; Pouchet C; Gilbert L; O’Brien PK; Serfas K; Broderick P; Houlston RS; Lesueur F; Bonora E; Muljo S; Schimke RN; Soglio DB-D; Arseneau J; Schultz KA; Priest JR; Nguyen V-H; Harach HR; Livingston DM; Foulkes WD; Tischkowitz M DICER1Mutations in Familial MultiNodular Goiter with and without Ovarian Sertoli-Leydig Cell Tumors. JAMA 2011, 305 (1), 68–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (99).Vedanayagam J; Chatila WK; Aksoy BA; Majumdar S; Skanderup AJ; Demir E; Schultz N; Sander C; Lai EC Cancer-Associated Mutations in DICER1 RNase IIIa and IIIb Domains Exert Similar Effects on MiRNA Biogenesis. Nat. Commun 2019, 10 (1), 3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (100).Wu MK; de Kock L; Conwell LS; Stewart CJR; King BR; Choong CS; Hussain K; Sabbaghian N; MacRae IJ; Fabian MR; Foulkes WD Functional Characterization of Multiple DICER1Mutations in an Adolescent. Endocrine-Related Cancer 2016, 23 (2), L1–L5. [DOI] [PubMed] [Google Scholar]
- (101).Oliver-Petit I; Bertozzi A-I; Grunenwald S; Gambart M; Pigeon-Kerchiche P; Sadoul J-L; Caron PJ; Savagner F Multinodular Goitre Is a Gateway for Molecular Testing of DICER1 Syndrome. Clinical Endocrinology 2019, 91 (5), 669–675. [DOI] [PubMed] [Google Scholar]
- (102).De Paolis E; Paragliola RM; Concolino P Spectrum of DICER1 Germline Pathogenic Variants in Ovarian Sertoli–Leydig Cell Tumor. J. Clin Med 2021, 10 (9), 1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (103).Heravi-Moussavi A; Anglesio MS; Cheng S-WG; Senz J; Yang W; Prentice L; Fejes AP; Chow C; Tone A; Kalloger SE; Hamel N; Roth A; Ha G; Wan ANC; Maines-Bandiera S; Salamanca C; Pasini B; Clarke BA; Lee AF; Lee C-H; Zhao C; Young RH; Aparicio SA; Sorensen PHB; Woo MMM; Boyd N; Jones SJM; Hirst M; Marra MA; Gilks B; Shah SP; Foulkes WD; Morin GB; Huntsman DG Recurrent Somatic DICER1Mutations in Nonepithelial Ovarian Cancers. New England Journal of Medicine 2012, 366 (3), 234–242. [DOI] [PubMed] [Google Scholar]
- (104).Kandasamy SK; Fukunaga R Phosphate-Binding Pocket in Dicer-2 PAZ Domain for High-Fidelity SiRNA Production. Proc. Natl. Acad. Sci. U. S. A 2016, 113 (49), 14031–14036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (105).Wojnicka M; Szczepanska A; Kurzynska-Kokorniak A Unknown Areas of Activity of Human Ribonuclease Dicer: A Putative Deoxyribonuclease Activity. Molecules 2020, 25 (6), 1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (106).Baronti L; Guzzetti I; Ebrahimi P; Friebe Sandoz S; Steiner E; Schlagnitweit J; Fromm B; Silva L; Fontana C; Chen AA; Petzold K Base-Pair Conformational Switch Modulates MiR-34a Targeting of Sirt1MRNA. Nature 2020, 583 (7814), 139–144. [DOI] [PubMed] [Google Scholar]
- (107).Baisden JT; Boyer JA; Zhao B; Hammond SM; Zhang Q Visualizing a Protonated RNA State That Modulates MicroRNA-21 Maturation. Nat. Chem. Biol 2021, 17 (1), 80–88. [DOI] [PubMed] [Google Scholar]
- (108).Kotar A; Ma S; Keane SC PH Dependence of C•A, G•A and A•A Mismatches in the Stem of Precursor MicroRNA-31. Biophys. Chem 2022, 283, 106763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (109).Taylor DW; Ma E; Shigematsu H; Cianfrocco MA; Noland CL; Nagayama K; Nogales E; Doudna JA; Wang HW Substrate-Specific Structural Rearrangements of Human Dicer. Nat. Struct Mol. Biol 2013, 20 (6), 662–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (110).Ramanathan A; Devarkar SC; Jiang F; Miller MT; Khan AG; Marcotrigiano J; Patel SS The Autoinhibitory CARD2-Hel2i Interface of RIG-I Governs RNA Selection. Nucleic Acids Res. 2016, 44 (2), 896–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (111).Brisse M; Ly H Comparative Structure and Function Analysis of the RIG-I-Like Receptors: RIG-1 and MDA5. Frontiers in Immunology 2019, 10, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (112).Li X; Lu C; Stewart M; Xu H; Strong RK; Igumenova T; Li P Structural Basis of Double-Stranded RNA Recognition by the RIG-I like Receptor MDA5. Arch. Biochem. Biophys 2009, 488 (1), 23–33. [DOI] [PubMed] [Google Scholar]
- (113).Kowalinski E; Lunardi T; McCarthy AA; Louber J; Brunel J; Grigorov B; Gerlier D; Cusack S Structural Basis for the Activation of Innate Immune Pattern-Recognition Receptor RIG-I by Viral RNA. Cell 2011, 147 (2), 423–435. [DOI] [PubMed] [Google Scholar]
- (114).Castellano L; Stebbing J Deep Sequencing of Small RNAs Identifies Canonical and Non-Canonical MiRNA and Endogenous SiRNAs in Mammalian Somatic Tissues. Nucleic Acids Res. 2013, 41 (5) , 3339–3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (115).Doyle M; Badertscher L; Jaskiewicz L; Güttinger S; Jurado S; Hugenschmidt T; Kutay U; Filipowicz W The Double-Stranded RNA Binding Domain of Human Dicer Functions as a Nuclear Localization Signal. RNA 2013, 19 (9), 1238–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (116).MacRae IJ; Zhou K; Doudna JA Structural Determinants of RNA Recognition and Cleavage by Dicer. Nat. Struct Mol. Biol 2007, 14 (10), 934–940. [DOI] [PubMed] [Google Scholar]
- (117).Sinkkonen L; Hugenschmidt T; Filipowicz W; Svoboda P Dicer Is Associated with Ribosomal DNA Chromatin in Mammalian Cells. PLoS One 2010, 5 (8), el2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (118).Wilson RCJ; Tambe A; Kidwell MA; Noland CL; Schneider CP; Doudna JA. Dicer-TRBP Complex Formation Ensures Accurate Mammalian MicroRNA Biogenesis. Mol. Cell 2015, 57 (3), 397–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (119).Rawling DC; Kohlway AS; Luo D; Ding SC; Pyle AM The RIG-I ATPase Core Has Evolved a Functional Requirement for Allosteric Stabilization by the Pincer Domain. Nucleic Acids Res. 2014, 42 (18), 11601–11611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (120).Wei X; Ke H; Wen A; Gao B; Shi J; Feng Y Structural Basis of MicroRNA Processing by Dicer-like 1. Nat. Plants 2021, 7 (10), 1389–1396. [DOI] [PubMed] [Google Scholar]
- (121).Dégut C; Schwarz V; Ponchon L; Barraud P; Micura R; Tisné C Design of Cross-Linked RNA/Protein Complexes for Structural Studies. Biochimie 2019, 164, 95–98. [DOI] [PubMed] [Google Scholar]
- (122).Du Z; Lee JK; Tjhen R; Stroud RM; James TL Structural and Biochemical Insights into the Dicing Mechanism of Mouse Dicer: A Conserved Lysine Is Critical for DsRNA Cleavage. Proc. Natl. Acad. Sci. U. S. A 2008, 105 (7), 2391–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]


