Abstract
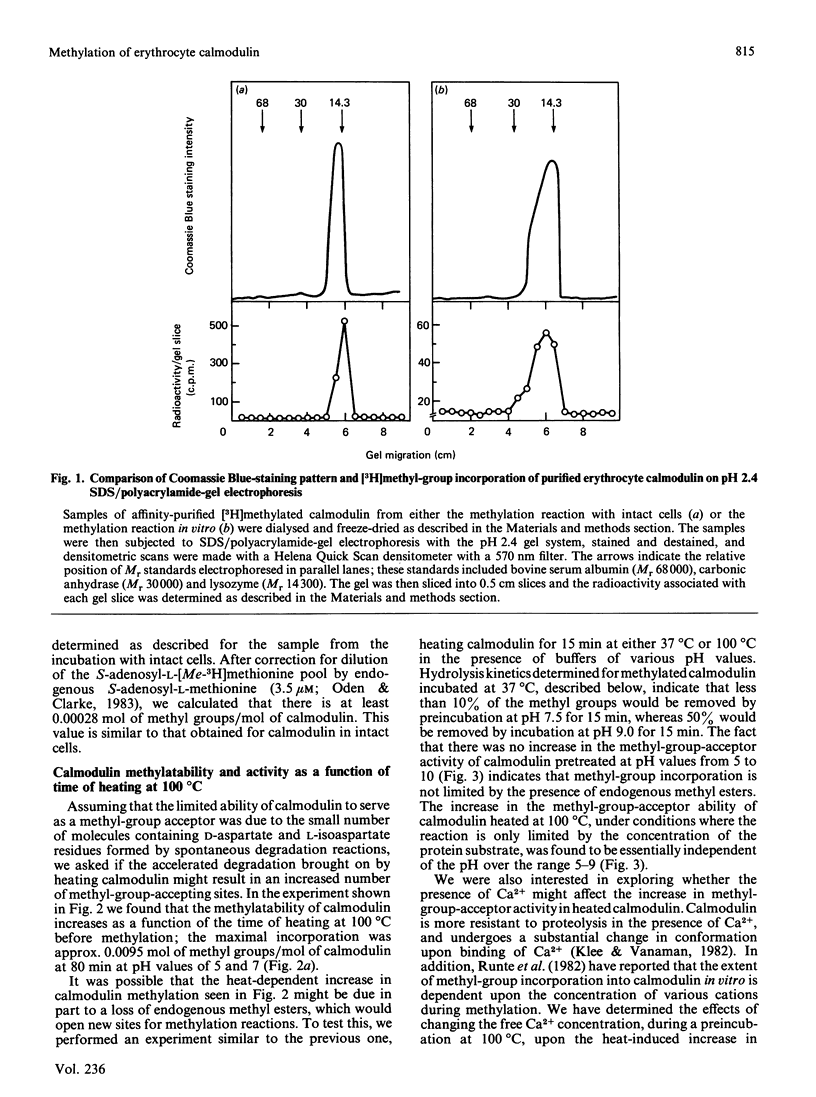
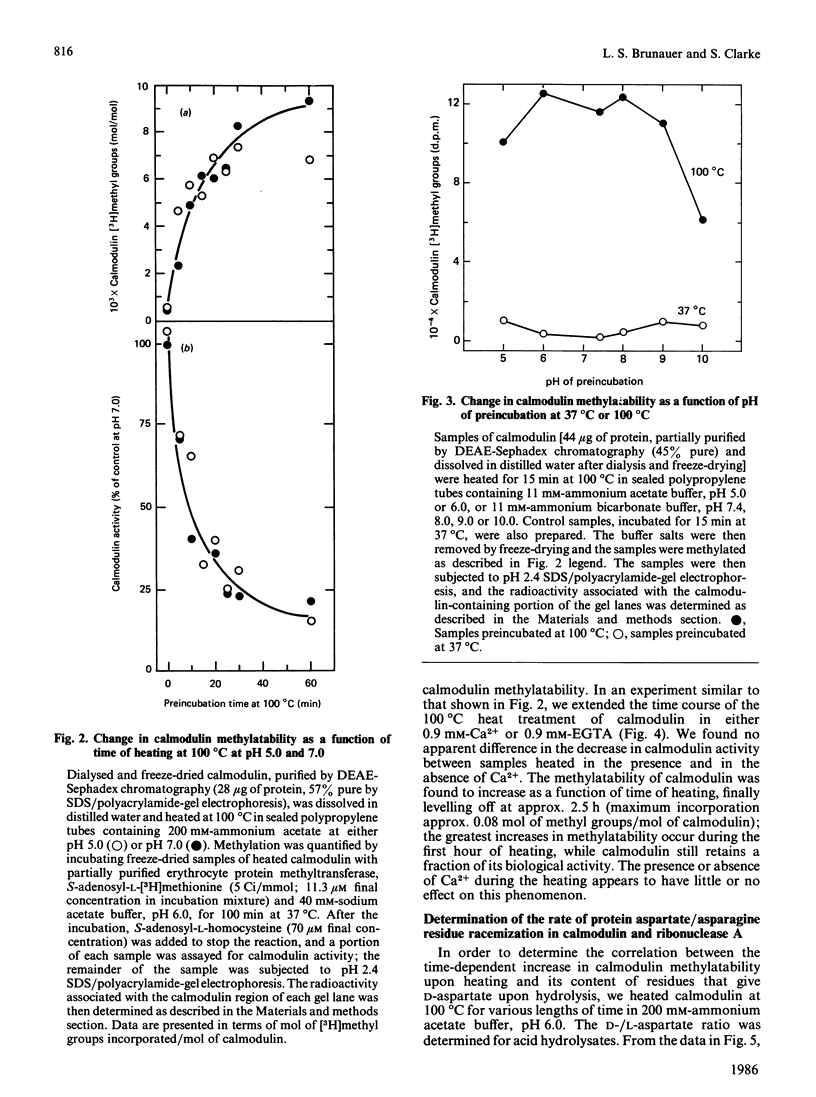
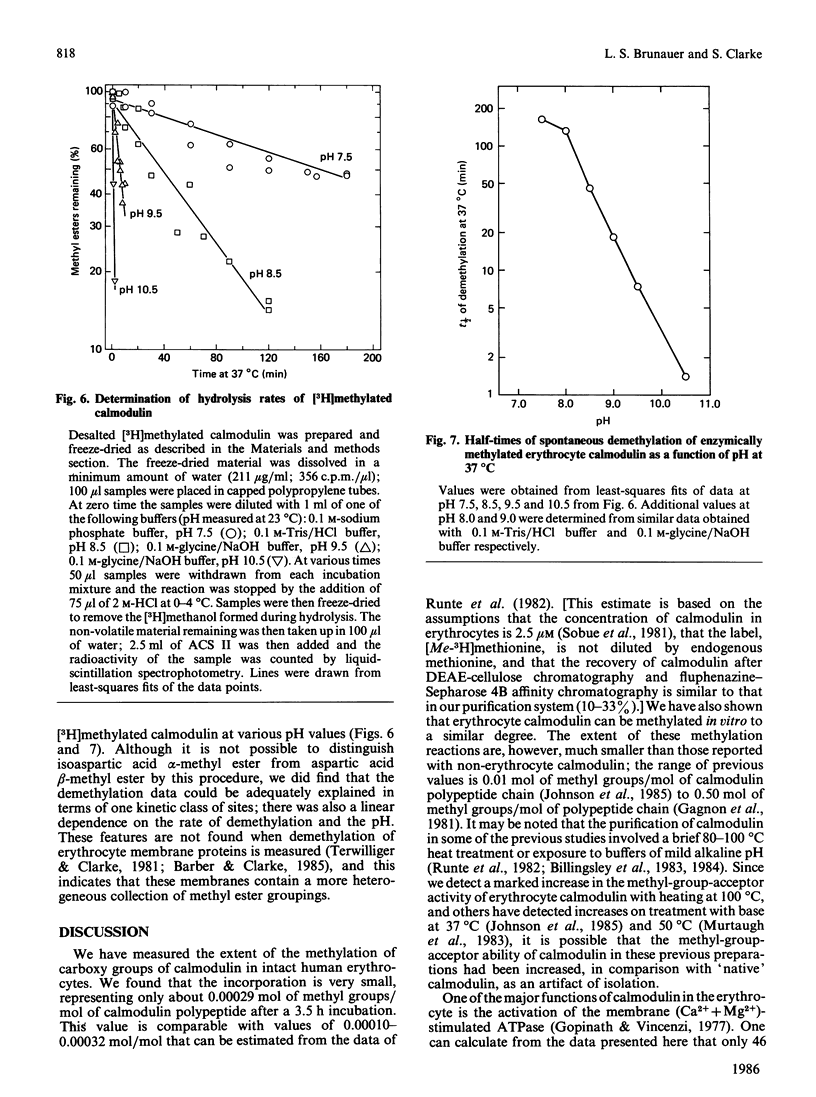
The physiological role of protein carboxy-group methylation reactions in human erythrocytes was studied with calmodulin as an endogenous methyl-group acceptor. The steady-state degree of calmodulin carboxy-group methylation is substoichiometric both in intact cells and in a lysed-cell system (about 0.0003 mol of methyl groups/mol of polypeptide). Purified erythrocyte calmodulin is a substrate for a partially purified erythrocyte carboxy-group methyltransferase and can be methylated to the extent of about 0.0007-0.001 mol of methyl groups/mol of polypeptide. This erythrocyte protein methyltransferase displays an apparent specificity for atypical racemized and/or isomerized D-aspartate and L-isoaspartate residues [McFadden & Clarke (1982) Proc. Natl. Acad. Sci. U.S.A. 79, 2460-2464; Murray & Clarke (1984) J. Biol. Chem. 259, 10722-10732]. Exposure of calmodulin to elevated temperatures before methylation results in racemization of aspartate and/or asparagine residues, and may result in isoaspartate formation as well. The methylatability of these samples also increases as a function of time of heating, independent of the pH (over the range pH 5-9) or Ca2+ concentration; the most significant increase occurs during the initial 60 min, when calmodulin retains a fraction of its biological activity. These results are consistent with the hypothesis that methylation of calmodulin may occur at these uncommon aspartate residues, but are not consistent with a regulatory role for the methylation reaction.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aswad D. W. Determination of D- and L-aspartate in amino acid mixtures by high-performance liquid chromatography after derivatization with a chiral adduct of o-phthaldialdehyde. Anal Biochem. 1984 Mar;137(2):405–409. doi: 10.1016/0003-2697(84)90106-4. [DOI] [PubMed] [Google Scholar]
- Aswad D. W. Stoichiometric methylation of porcine adrenocorticotropin by protein carboxyl methyltransferase requires deamidation of asparagine 25. Evidence for methylation at the alpha-carboxyl group of atypical L-isoaspartyl residues. J Biol Chem. 1984 Sep 10;259(17):10714–10721. [PubMed] [Google Scholar]
- Babu Y. S., Sack J. S., Greenhough T. J., Bugg C. E., Means A. R., Cook W. J. Three-dimensional structure of calmodulin. Nature. 1985 May 2;315(6014):37–40. doi: 10.1038/315037a0. [DOI] [PubMed] [Google Scholar]
- Barber J. R., Clarke S. Demethylation of protein carboxyl methyl esters: a nonenzymatic process in human erythrocytes? Biochemistry. 1985 Aug 27;24(18):4867–4871. doi: 10.1021/bi00339a021. [DOI] [PubMed] [Google Scholar]
- Barber J. R., Clarke S. Membrane protein carboxyl methylation increases with human erythrocyte age. Evidence for an increase in the number of methylatable sites. J Biol Chem. 1983 Jan 25;258(2):1189–1196. [PubMed] [Google Scholar]
- Bennett V. The membrane skeleton of human erythrocytes and its implications for more complex cells. Annu Rev Biochem. 1985;54:273–304. doi: 10.1146/annurev.bi.54.070185.001421. [DOI] [PubMed] [Google Scholar]
- Billingsley M. L., Velletri P. A., Roth R. H., DeLorenzo R. J. Carboxylmethylation of calmodulin inhibits calmodulin-dependent phosphorylation in rat brain membranes and cytosol. J Biol Chem. 1983 May 10;258(9):5352–5357. [PubMed] [Google Scholar]
- Billingsley M., Kuhn D., Velletri P. A., Kincaid R., Lovenberg W. Carboxylmethylation of phosphodiesterase attenuates its activation by ca2+-calmodulin. J Biol Chem. 1984 May 25;259(10):6630–6635. [PubMed] [Google Scholar]
- Bodanszky M., Kwei J. Z. Side reactions in peptide synthesis. VII. Sequence dependence in the formation of aminosuccinyl derivatives from beta-benzyl-aspartyl peptides. Int J Pept Protein Res. 1978 Aug;12(2):69–74. [PubMed] [Google Scholar]
- Bornstein P. Structure of alpha-1-CB8, a large cyanogen bromide produced fragment from the alpha-1 chain of rat collagen. The nature of a hydroxylamine-sensitive bond and composition of tryptic peptides. Biochemistry. 1970 Jun 9;9(12):2408–2421. doi: 10.1021/bi00814a004. [DOI] [PubMed] [Google Scholar]
- Burgess W. H., Howlett A. C., Kretsinger R. H., Gilman A. G. S49 lymphoma wild type and variant clones contain normal calcium dependent regulator. J Cyclic Nucleotide Res. 1978 Jun;4(3):175–181. [PubMed] [Google Scholar]
- Cheung W. Y. Calmodulin plays a pivotal role in cellular regulation. Science. 1980 Jan 4;207(4426):19–27. doi: 10.1126/science.6243188. [DOI] [PubMed] [Google Scholar]
- Clarke S., McFadden P. N., O'Connor C. M., Lou L. L. Isolation of D-aspartic acid beta-methyl ester from erythrocyte carboxyl methylated proteins. Methods Enzymol. 1984;106:330–344. doi: 10.1016/0076-6879(84)06033-x. [DOI] [PubMed] [Google Scholar]
- Clarke S. Protein carboxyl methyltransferases: two distinct classes of enzymes. Annu Rev Biochem. 1985;54:479–506. doi: 10.1146/annurev.bi.54.070185.002403. [DOI] [PubMed] [Google Scholar]
- Drickamer L. K. The red cell membrane contains three different adenosine triphophatases. J Biol Chem. 1975 Mar 10;250(5):1952–1954. [PubMed] [Google Scholar]
- Fairbanks G., Avruch J. Four gel systems for electrophoretic fractionation of membrane proteins using ionic detergents. J Supramol Struct. 1972;1(1):66–75. doi: 10.1002/jss.400010110. [DOI] [PubMed] [Google Scholar]
- Fairbanks G., Steck T. L., Wallach D. F. Electrophoretic analysis of the major polypeptides of the human erythrocyte membrane. Biochemistry. 1971 Jun 22;10(13):2606–2617. doi: 10.1021/bi00789a030. [DOI] [PubMed] [Google Scholar]
- Gagnon C., Kelly S., Manganiello V., Vaughan M., Odya C., Strittmatter W., Hoffman A., Hirata F. Modification of calmodulin function by enzymatic carboxylic methylation. Nature. 1981 Jun 11;291(5815):515–516. doi: 10.1038/291515a0. [DOI] [PubMed] [Google Scholar]
- Gopinath R. M., Vincenzi F. F. Phosphodiesterase protein activator mimics red blood cell cytoplasmic activator of (Ca2+-Mg2+)ATPase. Biochem Biophys Res Commun. 1977 Aug 22;77(4):1203–1209. doi: 10.1016/s0006-291x(77)80107-1. [DOI] [PubMed] [Google Scholar]
- Jackson R. L., Dedman J. R., Schreiber W. E., Bhatnagar P. K., Knapp R. D., Means A. R. Identification of epsilon-N-trimethyllysine in a rat testis calcium-dependent regulatory protein of cyclic nucleotide phosphodiesterase. Biochem Biophys Res Commun. 1977 Jul 25;77(2):723–729. doi: 10.1016/s0006-291x(77)80038-7. [DOI] [PubMed] [Google Scholar]
- Jamieson G. A., Jr, Vanaman T. C. Calcium-dependent affinity chromatography of calmodulin on an immobilized phenothiazine. Biochem Biophys Res Commun. 1979 Oct 12;90(3):1048–1056. doi: 10.1016/0006-291x(79)91932-6. [DOI] [PubMed] [Google Scholar]
- Jarrett H. W., Kyte J. Human erythrocyte calmodulin. Further chemical characterization and the site of its interaction with the membrane. J Biol Chem. 1979 Sep 10;254(17):8237–8244. [PubMed] [Google Scholar]
- Johnson B. A., Aswad D. W. Enzymatic protein carboxyl methylation at physiological pH: cyclic imide formation explains rapid methyl turnover. Biochemistry. 1985 May 7;24(10):2581–2586. doi: 10.1021/bi00331a028. [DOI] [PubMed] [Google Scholar]
- Johnson B. A., Freitag N. E., Aswad D. W. Protein carboxyl methyltransferase selectively modifies an atypical form of calmodulin. Evidence for methylation at deamidated asparagine residues. J Biol Chem. 1985 Sep 15;260(20):10913–10916. [PubMed] [Google Scholar]
- Kim S., Nochumson S., Chin W., Paik W. K. A rapid method for the purification of S-adenosylmethionine: protein-carboxyl O-methyltransferase by affinity chromatography. Anal Biochem. 1978 Feb;84(2):415–422. doi: 10.1016/0003-2697(78)90059-3. [DOI] [PubMed] [Google Scholar]
- Klee C. B., Vanaman T. C. Calmodulin. Adv Protein Chem. 1982;35:213–321. doi: 10.1016/s0065-3233(08)60470-2. [DOI] [PubMed] [Google Scholar]
- Kloog Y., Flynn D., Hoffman A. R., Axelrod J. Enzymatic carboxymethylation of the nicotinic acetylcholine receptor. Biochem Biophys Res Commun. 1980 Dec 31;97(4):1474–1480. doi: 10.1016/s0006-291x(80)80031-3. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Manalan A. S., Klee C. B. Calmodulin. Adv Cyclic Nucleotide Protein Phosphorylation Res. 1984;18:227–278. [PubMed] [Google Scholar]
- McFadden P. N., Clarke S. Methylation at D-aspartyl residues in erythrocytes: possible step in the repair of aged membrane proteins. Proc Natl Acad Sci U S A. 1982 Apr;79(8):2460–2464. doi: 10.1073/pnas.79.8.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray E. D., Jr, Clarke S. Metabolism of a synthetic L-isoaspartyl-containing hexapeptide in erythrocyte extracts. Enzymatic methyl esterification is followed by nonenzymatic succinimide formation. J Biol Chem. 1986 Jan 5;261(1):306–312. [PubMed] [Google Scholar]
- Murray E. D., Jr, Clarke S. Synthetic peptide substrates for the erythrocyte protein carboxyl methyltransferase. Detection of a new site of methylation at isomerized L-aspartyl residues. J Biol Chem. 1984 Sep 10;259(17):10722–10732. [PubMed] [Google Scholar]
- Murtaugh T. J., Rowe P. M., Vincent P. L., Wright L. S., Siegel F. L. Posttranslational modification of calmodulin. Methods Enzymol. 1983;102:158–170. doi: 10.1016/s0076-6879(83)02017-0. [DOI] [PubMed] [Google Scholar]
- O'Connor C. M., Aswad D. W., Clarke S. Mammalian brain and erythrocyte carboxyl methyltransferases are similar enzymes that recognize both D-aspartyl and L-isoaspartyl residues in structurally altered protein substrates. Proc Natl Acad Sci U S A. 1984 Dec;81(24):7757–7761. doi: 10.1073/pnas.81.24.7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor C. M., Clarke S. Analysis of erythrocyte protein methyl esters by two-dimensional gel electrophoresis under acidic separating conditions. Anal Biochem. 1985 Jul;148(1):79–86. doi: 10.1016/0003-2697(85)90630-x. [DOI] [PubMed] [Google Scholar]
- O'Connor C. M., Clarke S. Carboxyl methylation of cytosolic proteins in intact human erythrocytes. Identification of numerous methyl-accepting proteins including hemoglobin and carbonic anhydrase. J Biol Chem. 1984 Feb 25;259(4):2570–2578. [PubMed] [Google Scholar]
- O'Connor C. M., Clarke S. Methylation of erythrocyte membrane proteins at extracellular and intracellular D-aspartyl sites in vitro. Saturation of intracellular sites in vivo. J Biol Chem. 1983 Jul 10;258(13):8485–8492. [PubMed] [Google Scholar]
- Oden K. L., Clarke S. S-adenosyl-L-methionine synthetase from human erythrocytes: role in the regulation of cellular S-adenosylmethionine levels. Biochemistry. 1983 Jun 7;22(12):2978–2986. doi: 10.1021/bi00281a030. [DOI] [PubMed] [Google Scholar]
- Ohnishi S. T., Gall R. S. Characterization of the catalyzed phosphate assay. Anal Biochem. 1978 Aug 1;88(2):347–356. doi: 10.1016/0003-2697(78)90432-3. [DOI] [PubMed] [Google Scholar]
- Runte L., Jürgensmeier H. L., Unger C., Söling H. D. Calmodulin carboxylmethyl ester formation in intact human red cells and modulation of this reaction by divalent cations in vitro. FEBS Lett. 1982 Oct 4;147(1):125–130. doi: 10.1016/0014-5793(82)81025-9. [DOI] [PubMed] [Google Scholar]
- Sobue K., Muramoto Y., Fujita M., Kakiuchi S. Calmodulin-binding protein of erythrocyte cytoskeleton. Biochem Biophys Res Commun. 1981 Jun 16;100(3):1063–1070. doi: 10.1016/0006-291x(81)91931-8. [DOI] [PubMed] [Google Scholar]
- Terwilliger T. C., Clarke S. Methylation of membrane proteins in human erythrocytes. Identification and characterization of polypeptides methylated in lysed cells. J Biol Chem. 1981 Mar 25;256(6):3067–3076. [PubMed] [Google Scholar]


