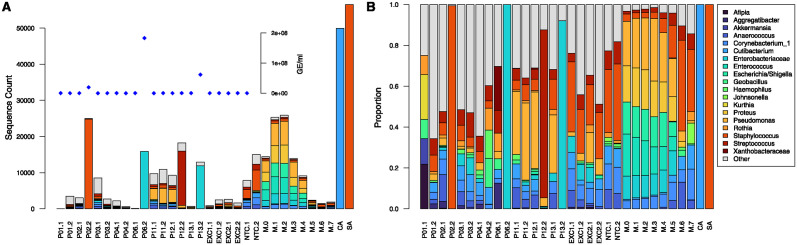
Fig 2. Taxonomic distribution of 16S RNA amplicons shown as raw counts (A) and proportions (B).
Patient labels are as in Table 1, with the number after the dot indicating previous surgery (1) or infection (2). Negative controls consist of extraction (EXC) and no-template (NTC) controls. Positive controls are the mock community (M.n), where n is the 10n dilution factor, and pure cultures of Cutibacterium acnes (CA) and Staphylococcus aureus (SA). Taxonomic categories that appeared in at least one sample at a proportion of 0.1 or more are given by name. The inset shows qPCR measurements of 16S RNA in genome equivalents (GE) per ml. Data for all 13 patients is shown in S1 Fig.