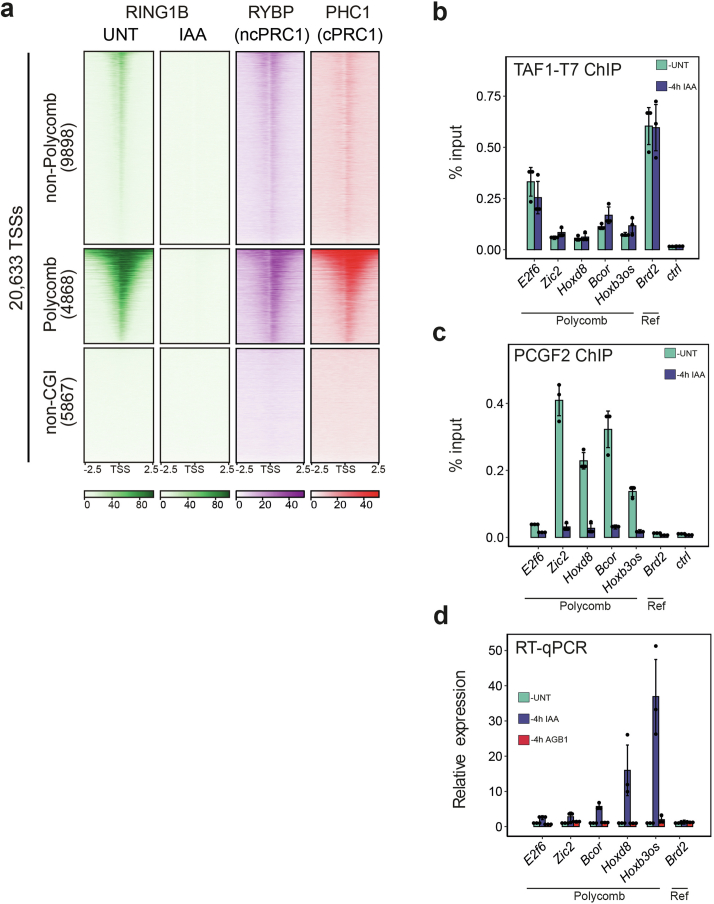
Extended Data Fig. 6. Genome-wide occupancy of canonical PRC1 complexes and their role in TFIID binding.
(a) Heat maps illustrating cChIP-seq signal for RINGB (all PRC1 complexes) (green, left), RYBP (ncPRC1, purple, middle), and PHC1 (cPRC1, red, right). TSSs were segregated into non-Polycomb (n = 9899), Polycomb (n = 4869), and non-CpG islands (n = 5869) groupings as indicated and ranked by decreasing RING1B signal. (b) ChIP-qPCR analysis of TAF1 chromatin occupancy at promoters of E2f6, Zic2, HoxD8, Bcor, Hoxb3os (Polycomb genes), as well as Brd2 (non-Polycomb gene, ‘Ref’) prior (UNT, dark blue) and after 4h depletion of PCGF2, a core component of cPRC1 (AGB1, light blue). Ctrl represents ChIP signal at a gene desert region. Error bars represent standard deviation from n = 3 biological replicates throughout the figure. (c) ChIP-qPCR analysis of PCGF2 as in (a), demonstrating its complete depletion from chromatin after 4h of treatment with AGB1. (d) Gene expression analysis of a panel of Polycomb genes using qRT-PCR after 4h depletion of RING1B (all PRC1 complexes, IAA) or PCGF2 (cPRC1 complexes, AGB1). Brd2 was used as a non-Polycomb gene (‘Ref’). Error bars represent standard deviation from n = 3 biological replicates. Source numerical data are available in source data.