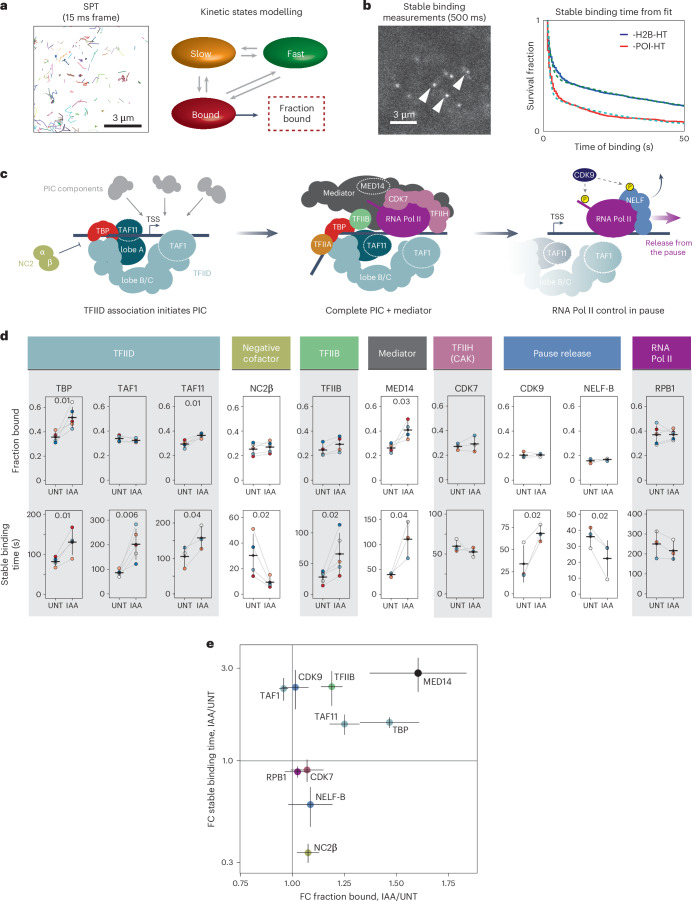
Fig. 4. PRC1 counteracts binding of early PIC-forming components.
a, An example of individually colour-coded single-molecule tracks acquired at high frame rate (left). These tracks are used for kinetic modelling in SPOT-ON42 to obtain bound fractions. b, An example frame from stable binding time measurements acquired at low frame rate with stably bound molecules indicated with arrow heads. Stable binding times for the protein of interest (POI) are extracted from bi-exponential fits (dotted lines) from cumulative distributions (solid lines) and corrected for photobleaching using estimates of stable binding of histone H2B-HT (blue). c, A cartoon illustrating stages of PIC assembly and transcription regulation. Protein factors studied by SPT are indicated. d, Dot plots illustrating the bound fractions (top) and stable binding time (bottom) for a panel of transcription regulators in untreated or PRC1-depleted (IAA-treated) conditions. Individually colour-coded dots represent values for individual biological replicates and are connected with grey lines, error bars represent s.d. and horizontal lines show the mean value. A minimum of three biological replicates were measured with approximately 100 cells per replicate for bound fraction analysis and approximately 20 cells for stable binding time measurements per biological replicate. P values were determined by one-sided paired t-tests and are presented whenever data reach statistical significance (P < 0.05). Bound fraction: TBP, P = 0.012903; TAF11, P = 0.01352; MED14, P = 0.032109; stable binding time: TBP, P = 0.010049; TAF1, P = 0.006401; TAF11, P = 0.040219; NC2β, P = 0.024727; TFIIB, P = 0.025326; MED14, P = 0.041231; CDK9, P = 0.023027; NELF-B, P = 0.024577. e, Scatter plot integrating the effects on bound fraction and stable biding times measured in SPT. Dots correspond to the mean fold change (FC) values for individual proteins and the error bars correspond to s.e.m. The data represents at least three biological replicates as indicated in d. Solid grey vertical and horizontal lines correspond to 1 (no change). Source numerical data are available in Source data.