Abstract
Coronaviruses (CoV), zoonotic viruses periodically emerging worldwide, represent a constant potential threat to humans. To date, seven human coronaviruses (HCoV) have been identified: HCoV-229E, HCoV-NL63, HCoV-OC43 and HCoV-HKU1, globally circulating in the human population (seasonal coronaviruses, sHCoV), and three highly-pathogenic coronaviruses, SARS-CoV, MERS-CoV and SARS-CoV-2. Although sHCoV generally cause only mild respiratory diseases, severe complications may occur in specific populations, highlighting the need for broad-spectrum anti-coronavirus drugs. Herein we show that indomethacin (INDO), a non-steroidal anti-inflammatory drug widely used in the clinic for its potent anti-inflammatory and analgesic properties, effectively inhibits the replication of Alpha-coronavirus HCoV-229E and Beta-coronavirus HCoV-OC43 in human lung-derived cells. Indomethacin does not interfere with HCoV binding or entry into target cells, but acts at late stages of the virus life cycle, inhibiting viral RNA synthesis and infectious viral particles production. Although INDO anti-inflammatory action is mediated by blocking cyclooxygenase-1 and -2 (COX-1/2) enzymatic activity, the antiviral effect appears to be cyclooxygenase-independent and is not mimicked by the potent COX-1/2 inhibitor aspirin. Interestingly we found that both seasonal HCoVs markedly (>100 fold) induce the expression of the pro-inflammatory mediator COX-2 in lung cells; notably, INDO-treatment was found to effectively inhibit virus-induced COX-2 expression at the transcriptional level, revealing an additional mechanism to prevent COX-2-mediated inflammatory reactions in HCoV-infected lung cells, besides COX activity inhibition. Altogether the results indicate that indomethacin, possessing both potent anti-inflammatory properties and a direct antiviral activity against HCoV, could be effective in the treatment of Alpha- and Beta-coronavirus infections.
Keywords: Antiviral, Cyclooxygenase-2, eIF2α, HCoV-229E, HCoV-OC43
1. Introduction
Coronaviruses (CoV), members of the family Coronaviridae, comprise a large number of enveloped, single-stranded positive-sense RNA viruses circulating among different natural hosts and causing respiratory, enteric, neurological and hepatic diseases of varying severity.1,2 Coronaviruses have the largest identified RNA genomes (typically ranging from 27 to 32 kb); the 5′-cap structure, along with a 3′-poly(A) tail, allows the CoV genome to act as an mRNA that can be translated by the host machinery.3 Despite their variety, CoVs share similarities in the organization of their genome: at the 5′ end, two large open reading frames (ORF, ORF1a and ORF1b) encode 15–16 non-structural proteins (nsp), of which 15 compose the viral replication and transcription complex containing RNA-processing and -modifying enzymes as well as an RNA proofreading function. ORFs that encode structural proteins [spike (S), envelope (E), membrane (M), nucleocapsid (N) and, in some CoV, hemagglutinin-esterase (HE)] and interspersed ORFs that encode accessory proteins are transcribed from the 3′ one-third of the genome.3 On the basis of their phylogenetic relationships and genomic structures, CoV are subdivided into four genera: Alpha-, Beta-, Gamma- and Delta-coronavirus; among these Alpha- and Beta-CoVs infect only mammals, whereas Gamma- and Delta-CoVs infect birds, and only occasionally can infect mammals.1,2 To date, seven human coronaviruses (HCoV) have been identified: HCoV-229E, HCoV-NL63, HCoV-OC43 and HCoV-HKU1, globally circulating in the human population (seasonal coronaviruses, sHCoV), and three highly pathogenic coronaviruses (HP-HCoV), SARS-CoV (Severe Acute Respiratory Syndrome-CoV), MERS-CoV (Middle-East Respiratory Syndrome-CoV) and the recently identified SARS-CoV-2.1,4 HP-HCoV infections can result in acute respiratory distress syndrome (ARDS), which may lead to long-term reduction in lung function and death1,5; in particular COVID-19, the disease associated with SARS-CoV-2, has caused over 775 million confirmed cases and 7 million deaths reported worldwide as of May 5, 2024 (https://covid19.who.int/).
The sHCoV generally cause mild respiratory tract infections and are estimated to contribute to 15–30 % of cases of common cold in humans.6,7 Although diseases are generally self-limiting, sHCoV can sometimes cause severe, life-threatening lower respiratory infections in a subset of patients, including elderly people, infants and immunocompromised patients.6, 7, 8, 9, 10, 11, 12, 13, 14, 15 Interestingly, whereas HCoV-229E was suggested to be involved in the development of Kawasaki disease, a rare condition that causes blood vessels inflammation,16,17 HCoV-OC43 has been shown to have neuroinvasive properties and to cause encephalitis in animal models.18,19 Moreover, both HCoV-OC43 and HCoV-229E were shown to establish persistent infections in cell cultures,20,21 while the presence of HCoV-OC43 RNA was detected in human brain autopsy samples from multiple sclerosis patients.8,19 These observations, together with the knowledge that HCoV infection does not induce long-lasting protective immunity,22 highlight the need for broad-spectrum anti-coronavirus drugs. So far, for SARS-CoV-2, two different RNA-dependent RNA polymerase inhibitors remdesivir23 and molnupiravir,24 and a viral protease inhibitor, Paxlovid (SARS-CoV-2 3CL protease inhibitor nirmatrelvir co-packaged with ritonavir), have been approved by health authorities in different countries.25 No specific antiviral drug or vaccine is presently available for seasonal coronavirus infections.
Indomethacin (INDO) is a non-steroidal anti-inflammatory drug (NSAID) widely used in the clinic for its potent anti-inflammatory and analgesic properties.26,27 INDO acts as a nonselective inhibitor of the bifunctional enzymes cyclooxygenase 1 and 2 (COX-1 and COX-2) that carry out two sequential reactions essential for the synthesis of prostaglandins and thromboxane.28 COX-1 is the constitutively expressed isoform; COX-2, although constitutively expressed in some tissues (e.g., brain and kidney), is primarily an inducible enzyme, whose expression is activated in response to cytokines, mitogens, endotoxin and hyperthermia in a variety of cell types.29,30
In addition to the anti-inflammatory/analgesic action, INDO has been known for several decades to also possess antiviral properties. Since the initial discovery,31 in-vitro and in-vivo studies have confirmed the antiviral activity of indomethacin against several human viral pathogens, including rotavirus,32 hepatitis B,33 herpesvirus,34 rhabdovirus35 and human immunodeficiency virus (HIV).36,37 We have previously shown that INDO is also effective against canine coronaviruses and the human coronavirus SARS-CoV;38 in addition, antiviral effects of INDO have been recently shown against SARS-CoV-2 in in-vitro models,39,40 as well as in clinical trials for the treatment of COVID-19 patients.41 Indomethacin ability to inhibit the replication of several unrelated DNA and RNA viruses points out to a host-mediated antiviral mechanism, rather than an effect on virus-specific enzymes. INDO was shown to selectively activate the double-stranded RNA-dependent protein kinase PKR, an interferon-induced serine-threonine protein kinase that plays an essential role in innate immunity response to viral infections, rapidly inducing the phosphorylation of the α-subunit of the eukaryotic initiation factor 2 (eIF2)42 and inhibiting viral protein synthesis.35 However, the mechanism at the basis of the antiviral activity of INDO remains largely unknown.
Herein we show that INDO effectively inhibits the replication of the Alpha-coronavirus HCoV-229E and the Beta-coronavirus HCoV-OC43 in human lung-derived cells, acting at late stages of the virus life cycle. Interestingly we found that both sHCoVs markedly induce the expression of the pro-inflammatory mediator COX-2 in lung cells, and that this effect is prevented by INDO treatment.
2. Materials and methods
2.1. Cell culture and treatments
Human normal lung MRC-5 fibroblasts (American Type Culture Collection, ATCC, CCL-171), embryonic kidney HEK293T cells (ATCC), and Caco-2 hACE2 cells, stably expressing the human angiotensin-converting enzyme 2 (hACE2) receptor, were grown at 37 °C in a 5 % CO2 atmosphere in EMEM (ATCC, MRC-5 cells) or DMEM (Euroclone, HEK293T and Caco-2 hACE2 cells) medium, supplemented with 10 % fetal calf serum (FCS), 2 mM glutamine and antibiotics. Generation of Caco-2 hACE2 cells has been described previously.43 Indomethacin and aspirin (ASA) were purchased from Sigma and dissolved into ethanol stock solutions. Compounds were diluted in culture medium, added to infected cells after the virus adsorption period and maintained in the medium for the duration of the experiment, unless differently specified. Controls received equal amounts of vehicle, which did not affect cell viability or virus replication. Cell viability was determined by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) to MTT formazan conversion assay (Sigma-Aldrich), as described.44 The 50 % lethal dose (LD50) was calculated using Prism 8.0 software (Graph-Pad Software Inc.). Microscopical examination of mock-infected or virus-infected cells was performed using a Leica DM-IL microscope and images were captured on a Leica DC 300 camera using Leica Image-Manager500 software.
2.2. Coronavirus infection and titration
Human coronaviruses HCoV-229E (ATCC) and HCoV-OC43 (ATCC) were used for this study. For virus infection, confluent MRC-5 cell monolayers were infected with HCoV for 1 h at 33 °C at a multiplicity of infection (MOI) of 0.01, 0.1 or 1 TCID50 (50 % tissue culture infectious dose)/cell. After the adsorption period, the viral inoculum was removed, and cell monolayers were washed three times with phosphate-buffered saline (PBS). Cells were maintained at 33 °C in growth medium containing 2 % FCS. Virus yield was determined at different times after infection by TCID50 infectivity assay, as described previously.45 The 50 % inhibitory concentration (IC50) of the compound tested was calculated using Prism 8.0 software.
2.3. Generation of SARS-CoV-2 S-pseudoviruses
SARS-CoV-2 S-pseudotyped lentiviral particles carrying a firefly luciferase reporter gene were generated as previously described.46 Briefly, HEK293T cells were co-transfected using Lipofectamine 2000 (Invitrogen, Thermo Scientific) with the pCAGGS-SARS2-S-G614 plasmid (Addgene plasmid #156421) encoding the SARS-CoV-2 spike carrying the D614G mutation, which increases virion spike infectivity, a gift from Hyeryun Choe and Michael Farzan,47 pCMVR 8.74 lentiviral packaging plasmid (a kind gift from R. Piva, University of Turin, Italy) and pLenti CMV Puro LUC plasmid (Addgene plasmid #17477), a gift from Eric Campeau and Paul Kaufman.48 At 48h after transfection, supernatants containing the viral particles were collected, titrated as described,46 and frozen at −80 °C until use.
2.4. Pseudovirus entry assay
Caco-2 hACE2 cells were plated at a density of 3 x 104 cells/well in white clear-bottom 96-well plates (Costar). After 24h, cells were infected with the pseudovirus suspension (50 μl: 5 x 105 RNA copies/well) for 4h. Luciferase activity (Relative Luciferase Units, RLU) was detected at 72h post-infection using Bright-Glo Luciferase Assay System Kit (Promega) in a Microplate Luminometer (Wallac-Perkin Elmer). All experiments were performed in triplicates.
2.5. Protein analysis and western blot
For analysis of proteins, whole-cell extracts (WCE) were prepared after lysis in High Salt Buffer (HSB) as described.49 For Western blot analysis, cell extracts (15 μg/sample) were separated by SDS-PAGE under reducing conditions and blotted to a nitrocellulose membrane. After blocking with 5 % skim milk solution, membranes were incubated with the selected antibodies, followed by incubation with peroxidase-labeled anti-rabbit or anti-mouse antibodies. Primary and secondary antibodies used are listed in Supplementary Table S1. Quantitative evaluation of proteins was determined as described.35,50 All results shown are representative of at least three independent experiments.
2.6. RNA extraction and real-time PCR analysis
Measurement of sHCoV genomic RNA was performed by quantitative Real-Time reverse-transcription PCR (qRT-PCR) analysis, as described.51 Briefly, total intracellular RNA from mock-infected or HCoV-infected cells was prepared using ReliaPrep RNA Cell Miniprep System (Promega) and reverse transcription was performed with PrimeScript RT Reagent Kit (Takara) according to the manufacturer's protocol. Extracellular viral RNA was extracted from 200 μl of the supernatant of sHCoV-infected cell cultures with Viral Nucleic Acid Extraction Kit II (Geneaid) and 10 μl aliquots were subjected to reverse transcription using SuperScript™ VILO™ cDNA Synthesis Kit (Life Technologies).52 qRT-PCR analysis was performed using specific primers listed in Supplementary Table S2, using SensiFAST SYBR® kit (Bioline) in a CFX96 Real-Time System (Bio-Rad). Primers specific for the membrane protein gene of HCoV-OC43 and HCoV-229E were previously described.53 Relative quantities of selected mRNAs were normalized to ribosomal L34 RNA levels in the same sample.52 All reactions were made in triplicate using samples derived from at least three biological repeats.
2.7. HCoV genomic RNA transfection
For HCoV genomic RNA transfection experiments, MRC-5 cell monolayers were infected with HCoV-229E or HCoV-OC43 for 1h at 33 °C at a MOI of 0.1 TCID50/cell and HCoV genomic RNA was extracted from the supernatants at 24h p.i. using TRIzol-LS reagent (Life Technologies), as described in the manufacturer's protocol. MRC-5 cells monolayers were mock-transfected or transfected with HCoV genomic RNA (1 μg/ml) using TransITmRNA Transfection Kit (Mirus Bio) at 33°C.52 After 4h, transfection medium was removed, and cells were treated with INDO or vehicle and maintained at 33 °C in growth medium containing 2 % FCS. After 48h, culture supernatants were collected for virus progeny titer determination, and cell monolayers were processed by Western blot analysis.
2.8. Immunofluorescence microscopy
MRC-5 cells grown in 8-well chamber slides (LabTek II) were infected with HCoV-229E or HCoV-OC43 and, after the adsorption period, were treated with INDO or vehicle for 24h. Cells were fixed, permeabilized and processed for immunofluorescence as described43 using selected antibodies, followed by decoration with Alexa Fluor 555- or 488-conjugated antibodies (Molecular Probes, Invitrogen). Primary and secondary antibodies used are listed in Supplementary Table S1. Nuclei were stained with Hoechst 33342 (Molecular Probes, Invitrogen). Images were captured using a ZEISS Axio Observer 7 inverted microscope with Apotome III and analyzed using ZEN 3.1 (blue edition) software. Images shown in all figures are representative of at least three random fields (scale-bars are indicated).
2.9. Statistical analysis
Statistical analyses were performed using Prism 8.0 software (GraphPad Software). Comparisons between two groups were made using Student's t-test; comparisons among groups were performed by one-way ANOVA with Bonferroni adjustments. p values ≤ 0.05 were considered significant. Data are expressed as the means ± standard deviations (S.D.) of results from duplicate or triplicate samples. Each experiment (in duplicate) was repeated at least twice.
3. Results and discussion
3.1. The non-steroidal anti-inflammatory drug indomethacin inhibits the replication of human seasonal coronaviruses HCoV-229E and HCoV-OC43 in in-vitro models
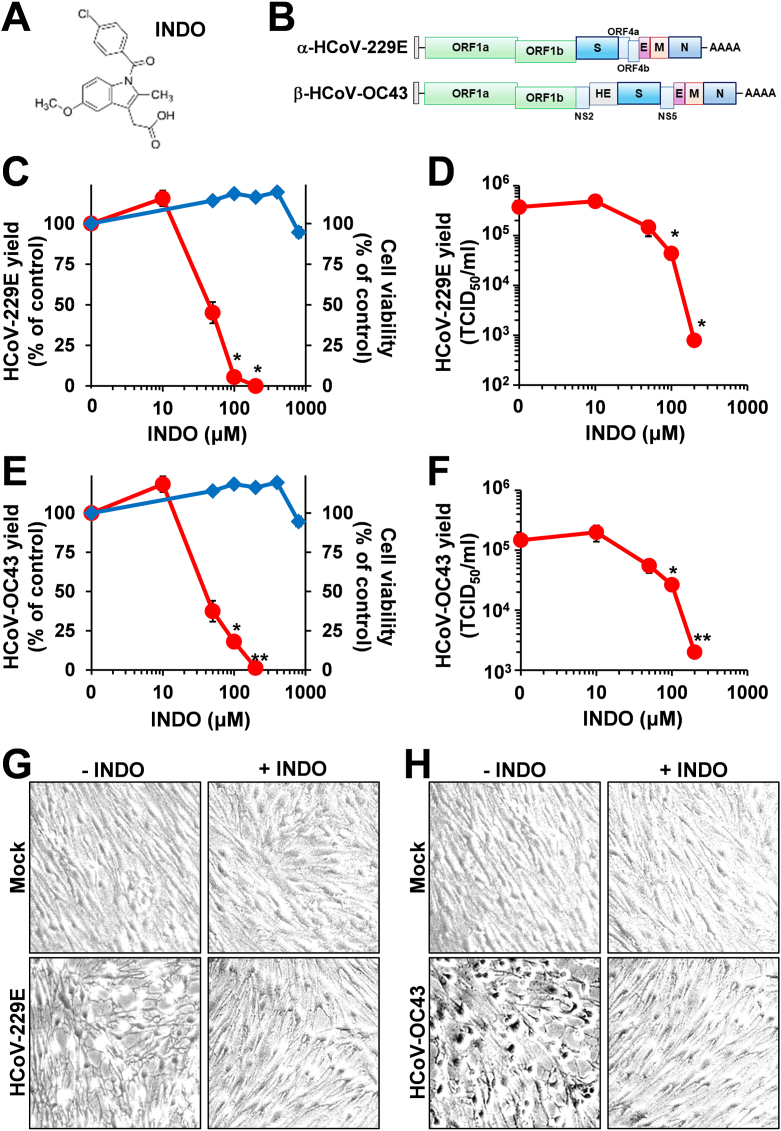
In order to investigate the antiviral activity of indomethacin (INDO, Fig. 1A) against sHCoV, MRC-5 cells derived from normal human lung tissue were infected with the Alpha-coronavirus HCoV-229E or the Beta-coronavirus HCoV-OC43 (Fig. 1B) at a MOI of 0.1 TCID50/cell and, after the 1h adsorption period, cells were treated with different concentrations of the drug. Controls received equal amounts of vehicle. At 24h post infection (p.i.), viral titers were determined in the supernatant of infected cells by TCID50 infectivity assay; in parallel, the effect of INDO on the viability of mock-infected MRC-5 cells was determined by MTT assay. INDO was found to possess a remarkable antiviral activity against HCoV-229E, reducing virus yield dose-dependently with an IC50 value of 45 μM (Fig. 1C and D), which is similar to the IC50 previously reported for INDO in SARS-CoV-infected cells.38 A greater than 99 % decrease in virus yield was detected at non-toxic concentrations (Fig. 1C and D). Similar results were obtained in MRC-5 cells infected with HCoV-OC43 (Fig. 1E and F). In addition to not being cytotoxic in MRC-5 cells at the effective antiviral concentrations with a CC50 > 800 μM (Fig. 1C and E), INDO (100 μM) was found to partially protect lung cells from the cytopathic effect caused by HCoV-229E and HCoV-OC43 infection for a period of 24h (Fig. 1G and H).
Fig. 1.
The non-steroidal anti-inflammatory drug indomethacin inhibits HCoV replication in human lung MRC-5 cells. (A) Structure of indomethacin (INDO). (B) Schematic representation of genome structure and classification of the human coronaviruses HCoV-229E and HCoV-OC43. ORF1a and ORF1b are represented as light green boxes; genes encoding structural proteins spike (S), nucleocapsid (N), envelope (E), membrane (M), and hemagglutinin-esterase (HE) and genes encoding accessory proteins are shown. (C–F) MRC-5 cell monolayers infected with HCoV-229E (C, D) or HCoV-OC43 (E, F) at an MOI of 0.1 TCID50/cell were treated with different concentrations of INDO or vehicle (0) after the adsorption period. Virus yield (O, red line) was determined at 24h p.i. by TCID50 infectivity assay. Data, expressed as percentage of control (C, E), represent the mean ± S.D. of triplicate samples. Data, expressed as TCID50/ml (D, F), represent the mean ± S.D. of duplicate samples from a representative experiment of three with similar results. ∗ = p < 0.05; ∗∗ = p < 0.01; ANOVA test. In parallel, cell viability (C, E, ◇, blue line) was determined by MTT assay in mock-infected cells. Absorbance (O.D.) of converted dye was measured at λ = 570 nm. Data, expressed as percentage of control, represent the mean ± S.D. of triplicate samples. (G, H) Cytoprotective effect of INDO (100 μM) in MRC-5 cells infected with HCoV-229E (G) or HCoV-OC43 (H) (0.1 TCID50/cell) at 24h p.i. (magnification: 100×). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Next, to determine whether INDO antiviral activity is affected by the number of HCoV infectious particles, MRC-5 cells were infected with HCoV-229E or HCoV-OC43 at different MOI (0.01, 0.1 or 1 TCID50/cell), and treated with 100 μM INDO or vehicle after the adsorption period; virus yield was determined at 24h p.i. by TCID50 infectivity assay. The results, shown in Fig. S1, demonstrate that INDO was effective under all conditions used, indicating that the drug inhibits sHCoV replication independently of the MOI.
As described in the Introduction, indomethacin exerts its anti-inflammatory effect by inhibiting cyclooxygenase-1 and cyclooxygenase-2 activity.27 We then investigated the effect of the potent COX-1/COX-2 inhibitor aspirin (ASA, Fig. S2A) on HCoV replication in human lung cells. MRC-5 cells were infected with HCoV-229E or HCoV-OC43 (0.1 TCID50/cell) and, after the adsorption period, were treated with different concentrations of ASA (1 μM–1 mM) or control vehicle. In parallel, ASA cytotoxicity was determined by MTT assay in mock-infected cells. At 24h p.i., HCoV-229E and HCoV-OC43 yields were determined by TCID50 infectivity assay. As previously shown in the case of different viruses,35,38 no antiviral activity against HCoV-229E or HCoV-OC43 was detected in ASA-treated cells up to concentrations as high as 1 mM (Fig. S2), suggesting that the anticoronavirus effect of INDO is independent of cyclooxygenase activity.
3.2. Indomethacin acts at post-entry level
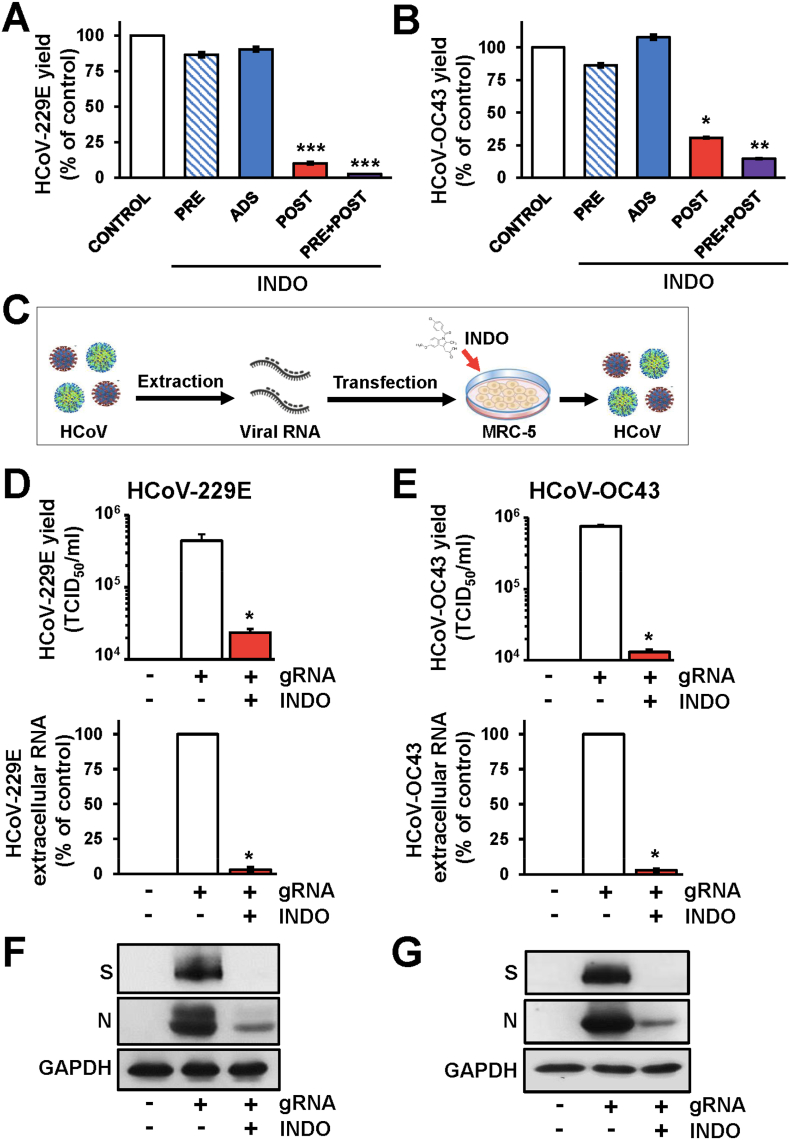
To determine the effect of treatment of lung cells with indomethacin before coronavirus infection, MRC-5 cells were treated with INDO (100 μM) or vehicle for 6h and the drug was removed before infection with HCoV-229E or HCoV-OC43 (0.1 TCID50/cell). In parallel, MRC-5 cells were infected with HCoV-229E or HCoV-OC43 in the absence of the drug and treated with INDO starting immediately after the 1h adsorption period for the duration of the experiment; alternatively, MRC-5 cells were treated for 6h before infection, and treatment was continued during and after the adsorption period. Finally to investigate whether INDO treatment affects HCoV receptor-binding or entry into host cells, MRC-5 cells were infected with HCoV-229E or HCoV-OC43 and treated with INDO or vehicle only during the 1h adsorption period, after which time the drug was removed. At 24h p.i. viral titers were determined in the supernatant of infected cells by TCID50 infectivity assay. As shown in Fig. 2A and B, INDO pretreatment did not affect sHCoV replication when the drug was removed before infection; treatment with the drug only during the virus adsorption period was also ineffective. On the other hand, treatment started after infection remarkably inhibited both HCoV-229E or HCoV-OC43 infectious particles production (Fig. 2A and B), suggesting that INDO acts at post-entry level.
Fig. 2.
Indomethacin acts at post-entry level. (A, B) MRC-5 cells infected with HCoV-229E (A) or HCoV-OC43 (B) (0.1 TCID50/cell) were treated with 100 μM INDO or vehicle (Control) at 6h before infection (PRE), only during the adsorption period (ADS) or immediately after the adsorption period in the absence (POST) or the presence of 6h pre-treatment (PRE + POST). Virus yield was determined at 24h p.i. by TCID50 infectivity assay. Data, expressed as percentage of control, represent the mean ± S.D. of duplicate samples from a representative experiment of two with similar results. ∗ = p < 0.05; ∗∗ = p < 0.01; ∗∗∗ = p < 0.001; ANOVA test. (C) Schematic representation of HCoV genomic RNA (gRNA) transfection. Created with BioRender.com. (D-G) MRC-5 cells were mock-transfected (-gRNA) or transfected with HCoV-229E (D, F) or HCoV-OC43 (E, G) gRNA for 4h and treated with 100 μM INDO (+) or vehicle (−) for 48h. Virus yield was determined at 48h after treatment by TCID50 assay (D, E, upper panels); in parallel, viral extracellular RNA levels were determined by qRT-PCR (D, E, lower panels). Data, expressed as TCID50/ml (D, E, upper panels) or percentage of control (D, E, lower panels) represent the mean ± S.D. of duplicate samples from a representative experiment of two with similar results. ∗ = p < 0.05; Student's t-test (D, E). In parallel samples levels of viral spike (S), viral nucleocapsid (N) and GAPDH proteins were determined by immunoblot analysis (IB) in HCoV-229E (F) or HCoV-OC43 (G) gRNA-transfected cells.
In order to rule out any effect of INDO on coronavirus adsorption, entry or uncoating, HCoV-229E or HCoV-OC43 genomic RNA was extracted from infectious virions and transfected into MRC-5 cell monolayers, as described in Materials and Methods, and schematically represented in Fig. 2C. After 4h, the transfection medium was removed and cells were treated with INDO (100 μM) or vehicle for 48h. At this time, viral titers were determined by TCID50 infectivity assay and extracellular HCoV genomic RNA levels were determined by qRT-PCR analysis in the supernatant of transfected cells; in parallel, levels of HCoV-229E and HCoV-OC43 spike (S) and nucleocapsid (N) proteins in transfected cells were analyzed in cell extracts by immunoblot. Both HCoV-229E or HCoV-OC43 genomic RNA transfection resulted in the production of infectious viral progeny at 48h after transfection (Fig. 2D and E). INDO treatment was found to greatly reduce the production of HCoV infectious particles and genomic extracellular RNA levels (Fig. 2D and E), as well as viral structural S and N proteins levels (Fig. 2F and G) in transfected cells. These results demonstrate that INDO does not interfere with the binding, entry or uncoating of HCoV-229E or HCoV-OC43 in target cells.
HCoV-229E and HCoV-OC43 are genetically dissimilar belonging to two distinct taxonomic genera (Alpha and Beta), and use different receptors: HCoV-229E has adopted the cell surface enzyme aminopeptidase-N (APN) as receptor, while HCoV-OC43 uses 9-O-acetylated sialic acid1, 2; the angiotensin-converting enzyme 2 (ACE2) is instead the receptor for HCoV-NL63, SARS-CoV-1 and SARS-CoV-2.1,4 To investigate whether indomethacin could interfere with SARS-CoV-2 entry into host cells, we engineered pseudotyped third-generation lentiviral particles to express the full-length SARS-CoV-2 S protein together with a luciferase (LUC) reporter to monitor infection. SARS-CoV-2 S-pseudotyped particles were generated in producer HEK293T cells and used to infect human colon Caco-2 cells stably expressing the human ACE2 receptor (Caco-2 hACE2)43 as described in Materials and Methods, and schematically represented in Fig. S3A. Caco-2 hACE2 cells were treated with INDO (100 μM) or vehicle for 2h and the drug was removed before infection with LUC-expressing SARS-CoV-2 S-pseudoviruses; alternatively, Caco-2 hACE2 cells were infected with SARS-CoV-2 S-pseudotyped particles and treated with 100 μM INDO or vehicle only during the 4h adsorption period, after which time the drug was removed. Pseudovirus entry was analyzed by measuring LUC activities 72h post-infection. Results in Fig. S3B show that drug treatment before or during adsorption had no effect on SARS-CoV-2 S-pseudoviruses infection, confirming that INDO does not affect SARS-CoV-2 S-pseudotyped particles entry.
Altogether, these results demonstrate that indomethacin does not interfere with the binding or entry of human coronaviruses into target cells, but acts at post-entry level.
3.3. Indomethacin acts at the late stages of the HCoV replication cycle
To investigate whether INDO affects an early step of the virus life cycle, MRC-5 cells were infected with HCoV-229E or HCoV-OC43 (0.1 TCID50/cell) and treated with 100 μM INDO starting immediately after the adsorption period or at different times after infection. At 24h p.i., virus yield was determined in the supernatant of infected cells by TCID50 infectivity assay. Interestingly, INDO treatment initiated between 0 and 6h after infection was equally effective in inhibiting HCoV replication, and it was still effective, although to a lesser extent, when treatment was started at 9h p.i.; the antiviral activity was instead impaired when treatment was started as late as 16h p.i. (Fig. S4). These results suggest that INDO does not affect early stages of the HCoV replication cycle.
We have previously shown that INDO causes a dysregulation of RNA and protein synthesis in cells infected with different viruses35,38: INDO was found to inhibit coronavirus RNA expression in a model of canine coronavirus (CCoV) infection38; on the other hand, we found that during infection with the negative-strand RNA virus VSV (Vesicular Stomatitis virus) INDO acts at the translational level, by rapidly inducing eIF2α phosphorylation via activation of the double-stranded RNA-activated protein kinase (PKR), and causing a block of viral protein expression.35,42
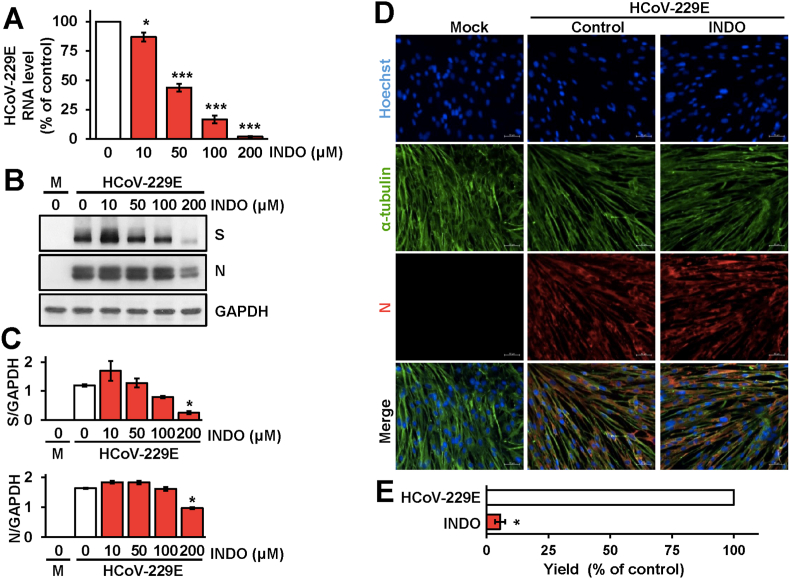
We therefore investigated the effect of the drug on viral RNA and structural protein expression during sHCoV infection. MRC-5 cells were mock-infected or infected with HCoV-229E (0.1 TCID50/cell) and treated with different concentrations of INDO starting immediately after the adsorption period. At 24h p.i., intracellular viral RNA levels were determined by qRT-PCR analysis; in parallel samples levels of the viral spike (S) and nucleocapsid (N) proteins were determined by immunoblot analysis using specific antibodies. As shown in Fig. 3A, INDO was found to decrease intracellular HCoV-229E RNA levels dose-dependently. Interestingly, differently from VSV infection, no significant differences in HCoV N and S protein levels were detected in INDO-treated cells, as compared to control, up to the concentration of 100 μM that caused a >90 % reduction in viral yield (Fig. 3B–E). Immunofluorescence microscopy analysis of MRC-5 cells infected with HCoV-229E (1 TCID50/cell) showed high levels of nucleocapsid protein in the cytoplasm of infected cells at 24h p.i. (Fig. 3D); comparable levels of N protein were detected in cells treated with INDO (100 μM) for 24h (Fig. 3D), confirming the immunoblot results.
Fig. 3.
Dose-dependent inhibition of HCoV-229E RNA and structural proteins expression by indomethacin. (A–C) MRC-5 cells mock-infected or infected with HCoV-229E (0.1 TCID50/cell) were treated with different concentrations of INDO or vehicle (0) after the adsorption period. Intracellular HCoV-229E RNA levels were determined by qRT-PCR at 24h p.i. (A). Relative quantities of HCoV-229E RNA were normalized to L34 RNA levels in the same sample. Data, expressed as percentage of untreated control, represent the mean ± S.D. of triplicate samples. ∗ = p < 0.05; ∗∗∗ = p < 0.001; ANOVA test. In parallel, levels of viral spike (S), viral nucleocapsid (N), and GAPDH proteins in HCoV-229E infected or mock-infected (M) cells were determined at 24h p.i. by IB (B). Relative amounts of S and N proteins were determined by densitometric analysis using ImageJ software, normalized to GAPDH in the same sample, and expressed as arbitrary units (C). Error bars indicate the mean ± S.D. of duplicate samples from a representative experiment of two with similar results. ∗ = p < 0.05; ANOVA test. (D) Immunofluorescence analysis of viral nucleocapsid protein (red) and α-tubulin (green) in MRC-5 cells mock-infected or infected with HCoV-229E (1 TCID50/cell) and treated with 100 μM INDO or vehicle for 24h. Nuclei are stained with Hoechst (blue). Merge images are shown. Scale bar, 50 μm. (E) MRC-5 cell monolayers infected with HCoV-229E (0.1 TCID50/cell) were treated with 100 μM INDO or vehicle after the adsorption period. Virus yield was determined at 24h p.i. by TCID50 infectivity assay. Data are expressed as percentage of control (n = 4). ∗ = p < 0.05; Student's t-test. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
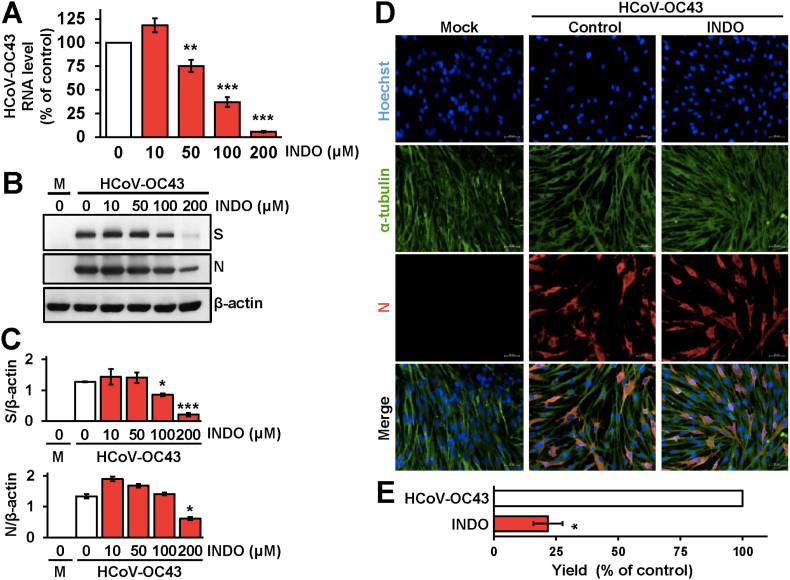
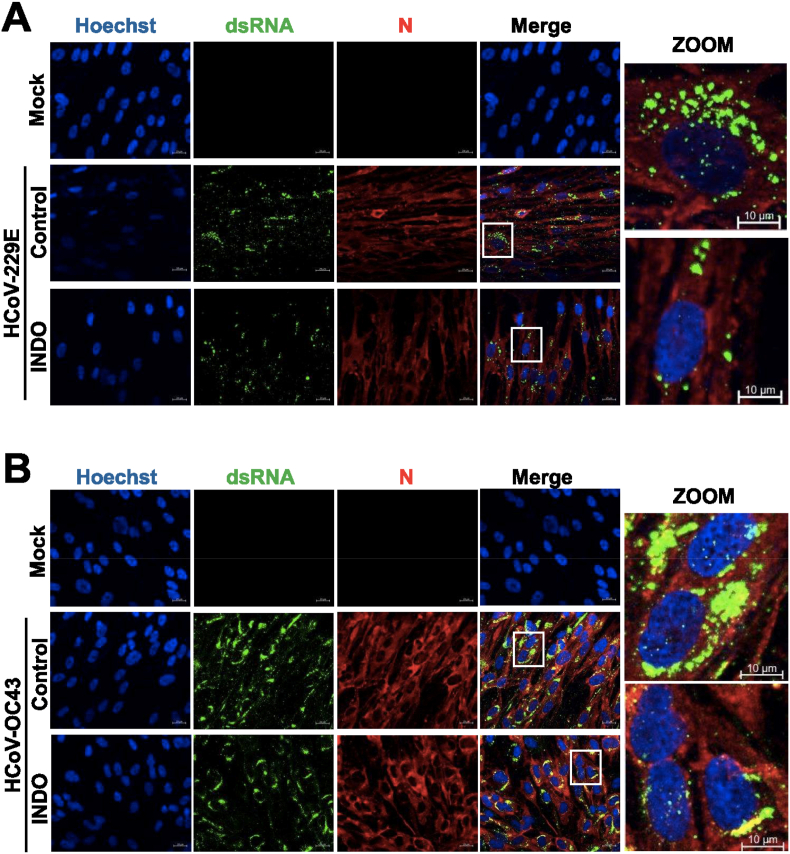
Similar results were obtained in a parallel set of experiments performed in MRC-5 cells infected with HCoV-OC43 (0.1 TCID50/cell) and treated with different concentrations of INDO starting immediately after the adsorption period. Also in this case INDO treatment caused a dose-dependent decrease in intracellular HCoV-OC43 RNA levels (Fig. 4A), while no significant differences in N protein levels were detected in INDO-treated cells up to the concentration of 100 μM despite the remarkable decrease in viral yield (Fig. 4B–E). Immunofluorescence microscopy studies also revealed that treatment with 100 μM INDO reduced the levels of both HCoV-229E and HCoV-OC43 double-strand RNA in lung cells without affecting viral nucleoprotein levels (Fig. 5). These results indicate that, differently from VSV-infected cells, the anticoronavirus activity of INDO is not primarily mediated by a translational block.
Fig. 4.
Effect of indomethacin on HCoV-OC43 RNA and structural proteins expression. (A–C) MRC-5 cells mock-infected or infected with HCoV-OC43 (0.1 TCID50/cell) were treated with different concentrations of INDO or vehicle (0) after the adsorption period. Intracellular HCoV-OC43 RNA levels were determined by qRT-PCR at 24h p.i. (A). Relative quantities of HCoV-OC43 RNA were normalized to L34 RNA levels in the same sample. Data, expressed as percentage of untreated control, represent the mean ± S.D. of triplicate samples. ∗∗ = p < 0.01; ∗∗∗ = p < 0.001; ANOVA test. In parallel, levels of viral spike (S), viral nucleocapsid (N), and β-actin proteins in HCoV-OC43 infected or mock-infected (M) cells were determined at 24h p.i. by IB (B). Relative amounts of S and N proteins were determined by densitometric analysis using ImageJ software, normalized to β-actin in the same sample, and expressed as arbitrary units (C). Error bars indicate the mean ± S.D. of duplicate samples from a representative experiment of two with similar results. ∗ = p < 0.05; ∗∗∗ = p < 0.001; ANOVA test. (D) Immunofluorescence analysis of viral nucleocapsid protein (red) and α-tubulin (green) in MRC-5 cells mock-infected or infected with HCoV-OC43 (1 TCID50/cell) and treated with 100 μM INDO or vehicle for 24h. Nuclei are stained with Hoechst (blue). Merge images are shown. Scale bar, 50 μm. (E) MRC-5 cell monolayers infected with HCoV-OC43 (0.1 TCID50/cell) were treated with 100 μM INDO or vehicle after the adsorption period. Virus yield was determined at 24h p.i. by TCID50 infectivity assay. Data are expressed as percentage of control (n = 4). ∗ = p < 0.05; Student's t-test. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Fig. 5.
Indomethacin treatment partially reduces HCoV dsRNA levels in lung cells. (A, B) Immunofluorescence analysis of viral dsRNA (green) and nucleocapsid protein (red) levels in MRC-5 cells mock-infected or infected with HCoV-229E (A) or HCoV-OC43 (B) (1 TCID50/cell) and treated with 100 μM INDO or vehicle (control) for 24h. Nuclei are stained with Hoechst (blue). Merge and zoom images are shown. Scale bar, 20 μm (zoom, 10 μm). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
As indicated above, INDO was shown to inhibit VSV protein synthesis by promoting the phosphorylation of eIF2α in monkey kidney cells.35 Notably, INDO treatment (100 μM) is able to induce eIF2α (Ser51) phosphorylation also in MRC-5 lung cells (Fig. S5). Phosphorylation of the factor occurs in a very rapid manner, starting as soon as 10 min after drug administration; however, this effect is transient, attenuating between 1 and 2h after treatment (Fig. S5A and B), and no differences in eIF2α phosphorylation levels were found in treated uninfected or sHCoV-infected cells at late times (16h) p.i. (Fig. S5D and F). The differential effects of INDO in sHCoV- vs. VSV-infected cells could be due to multiple factors: differently from VSV, whose replication cycle is very rapid (2–6 h)54 and is reliant on efficient cap-dependent mRNA translation in the early phase of infection,35 HCoV-229E and HCoV-OC43 are characterized by a much slower replication kinetics55 and may overcome a temporary translational block, due to the transient nature of the INDO-mediated eIF2α phosphorylation; moreover, the role of eIF2α at different stages of HCoV infection remains to be elucidated. Whereas the complex mechanisms underlying sHCoV-mediated control of the host translational apparatus are still poorly understood,56 it should be noted that HCoV-OC43 was recently shown to actively inhibit eIF2α phosphorylation in response to sodium arsenite.57,58 On the other hand, in the case of SARS-CoV infection it has been shown that SARS-CoV activates PKR, leading to sustained eIF2α phosphorylation that did not affect virus replication,59 suggesting that SARS-CoV is able to overcome the inhibitory effects of phosphorylated eIF2α on viral mRNA translation.
Whereas the mechanisms at the basis of the antiviral activity of indomethacin remain to be further elucidated, the presence of high levels of HCoV structural proteins in infected cells indicate that the drug is not affecting early stages of the replication cycle, including positive-sense RNA-genome translation into the pp1a and pp1ab polyproteins, their processing into different non-structural proteins and formation of the RNA replicase–transcriptase complex (RTC), nor RTC-driven generation of subgenomic RNAs, but could interfere with genomic RNA production and/or assembly of infectious viral particles in the host cell.
3.4. Indomethacin inhibits HCoV-induced cyclooxygenase-2 expression in lung cells
As indicated above, indomethacin is widely used in the clinic for its potent anti-inflammatory properties mainly due to inhibition of COX-2 enzymatic activity.26,27 Interestingly, it was previously reported that both SARS-CoV and SARS-CoV-2 infection is able to induce COX-2 expression in diverse systems in-vitro and in-vivo, and this effect has been linked to a virus-generated hyperinflammatory state60, 61, 62; however, little is known on the effect of seasonal coronavirus infection on COX-2 expression. We therefore investigated the effect of HCoV-OC43 and HCoV-229E infection on the expression of this critical inflammatory mediator in lung cells.
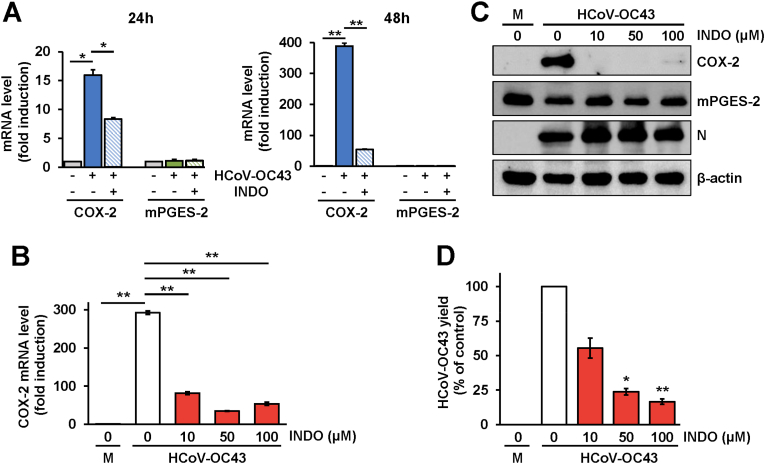
In a first set of experiments MRC-5 cells were mock-infected or infected with HCoV-OC43 (0.1 TCID50/cell), and treated with INDO (100 μM) or vehicle starting immediately after the adsorption period. At 24h and 48h p.i. COX-2 mRNA levels were determined by qRT-PCR analysis. Levels of microsomal prostaglandin E2 synthase (mPGES-2, an enzyme involved in prostaglandin E2 generation) mRNA were determined as control. HCoV-OC43 infection caused a marked increase (>15-fold) in COX-2 mRNA levels in lung cells already at 24h p.i.; notably COX-2 mRNA levels were found to increase more than 300-fold at 48h p.i. as compared to the mock-infected cells (Fig. 6A). No difference in mPGES-2 mRNA levels in the same samples was instead detected at both times p.i. (Fig. 6A). Indomethacin was found to greatly decrease the virus-induced COX-2 mRNA expression at both 24 and 48h after treatment, while it did not affect mPGES-2 mRNA levels (Fig. 6A).
Fig. 6.
HCoV-OC43 is a potent inducer of cyclooxygenase-2 expression in human lung cells: an effect inhibited by indomethacin. (A) MRC-5 cells were mock-infected or infected with HCoV-OC43 (0.1 TCID50/cell) and treated with 100 μM INDO or vehicle; at 24h or 48h p.i. levels of COX-2 and mPGES-2 mRNA were determined by qRT-PCR. Fold induction was calculated by comparing the increase of COX-2 and mPGES-2 in each sample to the mock-infected control, which was arbitrarily set to 1. Error bars indicate the mean ± S.D. of triplicate samples. ∗ = p < 0.05; ∗∗ = p < 0.01; Student's t-test. (B–D) MRC-5 cell monolayers mock-infected (M) or infected with HCoV-OC43 were treated with different concentrations of INDO or vehicle (0) after the adsorption period. Levels of COX-2 mRNA were determined by qRT-PCR at 48h p.i. (B). Fold induction was calculated by comparing the increase of COX-2 in each sample to the mock-infected control, which was arbitrarily set to 1. Error bars indicate the mean ± S.D. of triplicate samples. ∗∗ = p < 0.01; ANOVA test. Levels of COX-2, mPGES-2, viral nucleocapsid (N) and β-actin proteins in HCoV-OC43 infected or mock-infected (M) cells were determined at 48h p.i. by IB (C). In parallel samples virus yield was determined at 48h p.i. by TCID50 infectivity assay (D). Data, expressed as percentage of control, represent the mean ± S.D. of duplicate samples from a representative experiment of two with similar results. ∗ = p < 0.05; ∗∗ = p < 0.01; ANOVA test.
Next, MRC-5 cells were infected with HCoV-OC43 and, after the 1h adsorption period, were treated with different concentrations of the drug. At 48h p.i. COX-2 mRNA levels were determined by qRT-PCR analysis; in parallel samples levels of COX-2, mPGES-2 and viral nucleocapsid proteins were determined in cell extracts by immunoblot analysis, while virus yield was determined in the supernatant of infected cells by TCID50 infectivity assay. Interestingly, as shown in Fig. 6B and C, HCoV-OC43 infection not only caused a marked increase in COX-2 mRNA levels (Fig. 6B), but also induced the expression of high levels of COX-2 protein in infected cells at this time (Fig. 6C). No difference in mPGES-2 protein levels were detected in infected cells as compared to mock-infected cells (Fig. 6C). INDO was very effective in inhibiting virus-induced COX-2 mRNA and protein expression at all concentrations tested (Fig. 6B and C). Notably, also at 48h p.i., INDO treatment did not affect HCoV-OC43 N protein expression at concentrations that greatly reduced infectious viral particles productions in the same samples (Fig. 6D), confirming the results obtained at 24h (Fig. 4).
Similar results were obtained in a parallel set of experiments performed in MRC-5 cells infected with the α-HCoV-229E (0.1 TCID50/cell) and treated with different concentrations of INDO starting immediately after the adsorption period. HCoV-229E infection also triggered COX-2 expression in lung cells although to a lesser extent than the β-HCoV-OC43, causing an increase in COX-2 mRNA levels of more than 9-fold at 24h p.i. and more than 100-fold at 48h p.i., without affecting mPGES-2 mRNA levels (Fig. S6). Also in this case indomethacin treatment was found to be very effective in inhibiting the virus-induced COX-2 expression (Fig. S6B).
4. Conclusions
As a consequence of the high genomic nucleotide substitution rates, recombination and zoonotic potential, Coronaviridae is recognized as one of the most rapidly evolving virus family that represent a great threat to human health. In addition to the highly pathogenic human coronaviruses SARS-CoV, MERS-CoV and SARS-CoV-2, also seasonal coronaviruses, which generally cause mild upper respiratory tract diseases, may sometimes cause severe, life-threatening diseases in a subset of patients.6, 7, 8, 9, 10, 11, 12, 13, 14, 15 On this basis broad-spectrum anti-coronavirus drugs are highly needed.
We now show that the non-steroidal anti-inflammatory drug indomethacin effectively inhibits the replication of the Alpha-coronavirus HCoV-229E and the Beta-coronavirus HCoV-OC43 in human lung-derived cells. Indomethacin does not interfere with the binding or entry of human coronaviruses into target cells, but acts at late stages of the virus life cycle, inhibiting viral RNA synthesis and production of infectious viral particles.
Although INDO anti-inflammatory action is mediated by blocking COX-1 and COX-2 enzymatic activity thus inhibiting pro-inflammatory prostaglandin synthesis, the antiviral effect appears to be cyclooxygenase-independent, since it occurs at concentrations higher than those needed for COX inhibition (10−8, 10−9 M); in addition, the antiviral activity cannot be mimicked by the potent COX-1/2 inhibitor aspirin, that had no effect on either HCoV-229E or HCoV-OC43 replication up to millimolar concentrations. Interestingly we found that, similarly to the highly-pathogenic SARS-CoV and SARS-CoV-2,60, 61, 62 both seasonal HCoVs markedly induce COX-2 expression in lung cells. Whereas the mechanism(s) involved in sHCoV-mediated COX-2 expression remains to be elucidated, we show that INDO treatment effectively inhibits virus-induced COX-2 mRNA transcription. These results indicate that indomethacin is able to inhibit coronavirus-induced COX-2-mediated inflammatory reactions by acting at two separate levels, reducing COX-2 levels in infected lung cells, in addition to inhibiting COX enzymatic activity.
Altogether the results described indicate that indomethacin, possessing both anti-inflammatory properties and a direct antiviral activity against HCoV, could be effective in the treatment of Alpha- and Beta-coronavirus infections.
Funding
This research was supported by: the Italian Ministry of University and Scientific Research (PRIN project N 2010PHT9NF-006); EU funding within the NextGeneration EU-MUR PNRR Extended Partnership initiative on Emerging Infectious Diseases (Project no. PE00000007, INF-ACT).
CRediT authorship contribution statement
Caterina Tramontozzi: Formal analysis, Investigation, Data curation, Validation, Writing–original draft. Anna Riccio: Investigation, Validation, Writing–original draft. Silvia Pauciullo: Methodology, Validation. Simone La Frazia: Investigation, Methodology, Validation. Antonio Rossi: Funding acquisition, Investigation, Methodology. M. Gabriella Santoro: Conceptualization, Formal analysis, Funding acquisition, Supervision, Writing-original draft, Writing-review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors thank Roberto Piva, University of Turin, Italy, for providing the pCMVR 8.74 lentiviral packaging plasmid; Caterina Tramontozzi is enrolled in the PhD Program in Cellular and Molecular Biology, Department of Biology, University of Rome Tor Vergata, Rome, Italy and is the recipient of a fellowship partially funded by Alfasigma S.p.A. (Bologna, Italy) and Regione Lazio.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jve.2024.100387.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fung T.S., Liu D.X. Human coronavirus: host-pathogen interaction. Annu Rev Microbiol. 2019;73:529–557. doi: 10.1146/annurev-micro-020518-115759. [DOI] [PubMed] [Google Scholar]
- 3.V'kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol. 2021;19:155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Markov P.V., Ghafari M., Beer M., et al. The evolution of SARS-CoV-2. Nat Rev Microbiol. 2023;21:361–379. doi: 10.1038/s41579-023-00878-2. [DOI] [PubMed] [Google Scholar]
- 5.Lamers M.M., Haagmans B.L. SARS-CoV-2 pathogenesis. Nat Rev Microbiol. 2022;20:270–284. doi: 10.1038/s41579-022-00713-0. [DOI] [PubMed] [Google Scholar]
- 6.Lim Y.X., Ng Y.L., Tam J.P., Liu D. Human coronaviruses: a review of virus–host interactions. Diseases. 2016;4:26. doi: 10.3390/diseases4030026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu D.X., Liang J.Q., Fung T.S. In: Encyclopedia of Virology. fourth ed. Bamford D., Zuckerman M., editors. Elsevier Ltd., Academic Press; 2021. Human coronavirus-229e, -OC43, -NL63, and -HKU1 (Coronaviridae) pp. 428–440. [DOI] [Google Scholar]
- 8.Arbour N., Day R., Newcombe J., Talbot P.J. Neuroinvasion by human respiratory coronaviruses. J Virol. 2000;74:8913–8921. doi: 10.1128/JVI.74.19.8913-8921.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiu S.S., Hung Chan K., Wing Chu K., et al. Human coronavirus NL63 infection and other coronavirus infections in children hospitalized with acute respiratory disease in Hong Kong, China. Clin Infect Dis. 2005;40:1721–1729. doi: 10.1086/430301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gorse G.J., O'Connor T.Z., Hall S.L., Vitale J.N., Nichol K.L. Human coronavirus and acute respiratory illness in older adults with chronic obstructive pulmonary disease. J Infect Dis. 2009;199:847–857. doi: 10.1086/597122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacomy H., Fragoso G., Almazan G., Mushynski W.E., Talbot P.J. Human coronavirus OC43 infection induces chronic encephalitis leading to disabilities in BALB/C mice. Virology. 2006;349:335–346. doi: 10.1016/j.virol.2006.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morfopoulou S., Brown J.R., Davies E.G., et al. Human coronavirus OC43 associated with fatal encephalitis. N Engl J Med. 2016;375:497–498. doi: 10.1056/NEJMc1509458. [DOI] [PubMed] [Google Scholar]
- 13.Pene F., Merlat A., Vabret A., et al. Coronavirus 229E-related pneumonia in immunocompromised patients. Clin Infect Dis. 2003;37:929–932. doi: 10.1086/377612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Risku M., Lappalainen S., Räsänen S., Vesikari T. Detection of human coronaviruses in children with acute gastroenteritis. J Clin Virol. 2010;48:27–30. doi: 10.1016/j.jcv.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Z., Liu W., Zhang S., et al. Two novel human coronavirus OC43 genotypes circulating in hospitalized children with pneumonia in China. Emerg Microb Infect. 2022;11:168–171. doi: 10.1080/22221751.2021.2019560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Esper F., Shapiro E.D., Weibel C., Ferguson D., Landry M.L., Kahn J.S. Association between a novel human coronavirus and Kawasaki disease. J Infect Dis. 2005;191:499–502. doi: 10.1086/428291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shirato K., Imada Y., Kawase M., Nakagaki K., Matsuyama S., Taguchi F. Possible involvement of infection with human coronavirus 229E, but not NL63, in Kawasaki disease. J Med Virol. 2014;86:2146–2153. doi: 10.1002/jmv.23950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bergmann C.C., Lane T.E., Stohlman S.A. Coronavirus infection of the central nervous system: host-virus stand-off. Nat Rev Microbiol. 2006;4:121–132. doi: 10.1038/nrmicro1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng Q., Yang Y., Gao J. Infectivity of human coronavirus in the brain. EBioMedicine. 2020;56 doi: 10.1016/j.ebiom.2020.102799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arbour N., Côté G., Lachance C., Tardieu M., Cashman N.R., Talbot P.J. Acute and persistent infection of human neural cell lines by human coronavirus OC43. J Virol. 1999;73:3338–3350. doi: 10.1128/JVI.73.4.3338-3350.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arbour N., Ekandé S., Côté G., et al. Persistent infection of human oligodendrocytic and neuroglial cell lines by human coronavirus 229E. J Virol. 1999;73:3326–3337. doi: 10.1128/JVI.73.4.3326-3337.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edridge A.W.D., Kaczorowska J., Hoste A.C.R., et al. Seasonal coronavirus protective immunity is short-lasting. Nat Med. 2020;26:1691–1693. doi: 10.1038/s41591-020-1083-1. [DOI] [PubMed] [Google Scholar]
- 23.Santoro M.G., Carafoli E. Remdesivir: from ebola to COVID-19. Biochem Biophys Res Commun. 2021;538:145–150. doi: 10.1016/j.bbrc.2020.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jayk Bernal A., Gomes da Silva M.M., Musungaie D.B., et al. MOVe-OUT Study Group Molnupiravir for oral treatment of Covid-19 in nonhospitalized patients. N Engl J Med. 2022;386:509–520. doi: 10.1056/NEJMoa2116044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hammond J., Leister-Tebbe H., Gardner A., et al. EPIC-HR Investigators Oral nirmatrelvir for high-risk, nonhospitalized adults with Covid-19. N Engl J Med. 2022;386:1397–1408. doi: 10.1056/NEJMoa2118542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vane J.R., Botting R.M. Mechanism of action of antiinflammatory drugs. Int J Tissue React. 1998;20:3–15. [PubMed] [Google Scholar]
- 27.Blobaum A.L., Marnett L.J. Structural and functional basis of cyclooxygenase inhibition. J Med Chem. 2007;50:1425–1441. doi: 10.1021/jm0613166. [DOI] [PubMed] [Google Scholar]
- 28.Vane J.R., Bakhle Y.S., Botting R.M. Cyclooxygenases 1 and 2. Annu Rev Pharmacol Toxicol. 1998;38:97–120. doi: 10.1146/annurev.pharmtox.38.1.97. [DOI] [PubMed] [Google Scholar]
- 29.Rouzer C.A., Marnett L.J. Cyclooxygenases: structural and functional insights. J Lipid Res. 2009;50:S29–S34. doi: 10.1194/jlr.R800042-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rossi A., Coccia M., Trotta E., Angelini M., Santoro M.G. Regulation of cyclooxygenase-2 expression by heat: a novel aspect of heat shock factor 1 function in human cells. PLoS One. 2012;7 doi: 10.1371/journal.pone.0031304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Inglot A.D. Comparison of the antiviral activity in vitro of some non-steroidal anti-inflammatory drugs. J Gen Virol. 1969;4:203–214. doi: 10.1099/0022-1317-4-2-203. [DOI] [PubMed] [Google Scholar]
- 32.Rossen J.W., Bouma J., Raatgeep R.H., Büller H.A., Einerhand A.W. Inhibition of cyclooxygenase activity reduces rotavirus infection at a postbinding step. J Virol. 2004;78:9721–9730. doi: 10.1128/JVI.78.18.9721-9730.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bahrami H., Daryani N.E., Haghpanah B., et al. Effects of indomethacin on viral replication markers in asymptomatic carriers of hepatitis B: a randomized, placebo-controlled trial. Am J Gastroenterol. 2005;100:856–861. doi: 10.1111/j.1572-0241.2005.41144.x. [DOI] [PubMed] [Google Scholar]
- 34.Zhu H., Cong J.P., Yu D., Bresnahan W.A., Shenk T.E. Inhibition of cyclooxygenase 2 blocks human cytomegalovirus replication. Proc Natl Acad Sci USA. 2002;99:3932–3937. doi: 10.1073/pnas.052713799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Amici C., La Frazia S., Brunelli C., Balsamo M., Angelini M., Santoro M.G. Inhibition of viral protein translation by indomethacin in vesicular stomatitis virus infection: role of eIF2α kinase PKR. Cell Microbiol. 2015;17:1391–1404. doi: 10.1111/cmi.12446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramirez J., Alcami J., Arnaiz-Villena A., et al. Indomethacin in the relief of AIDS symptoms. Lancet. 1986;2:570. doi: 10.1016/s0140-6736(86)90132-7. [DOI] [PubMed] [Google Scholar]
- 37.Bourinbaiar A.S., Lee-Huang S. Potentiation of anti-HIV activity of anti-inflammatory drugs, dexamethasone and indomethacin, by MAP30, the antiviral agent from bitter melon. Biochem Biophys Res Commun. 1995;208:779–785. doi: 10.1006/bbrc.1995.1405. [DOI] [PubMed] [Google Scholar]
- 38.Amici C., Di Caro A., Ciucci A., et al. Indomethacin has a potent antiviral activity against SARS coronavirus. Antivir Ther. 2006;11:1021–1030. doi: 10.1177/135965350601100803. [DOI] [PubMed] [Google Scholar]
- 39.Abdel Shaheed C., Beardsley J., Day R.O., McLachlan A.J. Immunomodulatory effects of pharmaceutical opioids and antipyretic analgesics: mechanisms and relevance to infection. Br J Clin Pharmacol. 2022;88:3114–3131. doi: 10.1111/bcp.15281. [DOI] [PubMed] [Google Scholar]
- 40.Wang Y., Li P., Xu L., et al. Combating pan-coronavirus infection by indomethacin through simultaneously inhibiting viral replication and inflammatory response. iScience. 2023;26 doi: 10.1016/j.isci.2023.107631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ravichandran R., Mohan S.K., Sukumaran S.K., et al. An open label randomized clinical trial of Indomethacin for mild and moderate hospitalised Covid-19 patients. Sci Rep. 2022;12 doi: 10.1038/s41598022103701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brunelli C., Amici C., Angelini M., Fracassi C., Belardo G., Santoro M.G. The non-steroidal anti-inflammatory drug indomethacin activates the eIF2α kinase PKR, causing a translational block in human colorectal cancer cells. Biochem J. 2012;443:379–386. doi: 10.1042/BJ20111236. [DOI] [PubMed] [Google Scholar]
- 43.Riccio A., Santopolo S., Rossi A., Piacentini S., Rossignol J.F., Santoro M.G. Impairment of SARS-CoV-2 spike glycoprotein maturation and fusion activity by nitazoxanide: an effect independent of spike variants emergence. Cell Mol Life Sci. 2022;79:227. doi: 10.1007/s00018-022-04246-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Piacentini S., La Frazia S., Riccio A., et al. Nitazoxanide inhibits paramyxovirus replication by targeting the fusion protein folding: role of glycoprotein-specific thiol oxidoreductase ERp57. Sci Rep. 2018;8 doi: 10.1038/s41598-018-28172-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Santoro M.G., Favalli C., Mastino A., Jaffe B.M., Esteban M., Garaci E. Antiviral activity of a synthetic analog of prostaglandin A in mice infected with influenza A virus. Arch Virol. 1988;99:89–100. doi: 10.1007/BF01311026. [DOI] [PubMed] [Google Scholar]
- 46.Minenkova O., Santapaola D., Milazzo F.M., et al. Human inhalable antibody fragments neutralizing SARS-CoV-2 variants for COVID-19 therapy. Mol Ther. 2022;30:1979–1993. doi: 10.1016/j.ymthe.2022.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang L., Jackson C.B., Mou H., et al. SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat Commun. 2020;11:6013. doi: 10.1038/s41467-020-19808-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Campeau E., Ruhl V.E., Rodier F., et al. A versatile viral system for expression and depletion of proteins in mammalian cells. PLoS One. 2009;4 doi: 10.1371/journal.pone.0006529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Santoro M.G., Jaffe B.M., Garaci E., Esteban M. Antiviral effect of prostaglandins of the A series: inhibition of Vaccinia virus replication in cultured cells. J Gen Virol. 1982;63:435–440. doi: 10.1099/0022-1317-63-2-435. [DOI] [PubMed] [Google Scholar]
- 50.Santoro M.G., Amici C., Elia G., Benedetto A., Garaci E. Inhibition of virus protein glycosylation as the mechanism of the antiviral action of prostaglandin A in Sendai virus-infected cells. J Gen Virol. 1989;70:789–800. doi: 10.1099/0022-1317-70-4-789. [DOI] [PubMed] [Google Scholar]
- 51.Coccia M., Rossi A., Riccio A., Trotta E., Santoro M.G. Human NF-κB repressing factor acts as a stress-regulated switch for ribosomal RNA processing and nucleolar homeostasis surveillance. Proc Natl Acad Sci USA. 2017;114:1045–1050. doi: 10.1073/pnas.1616112114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Piacentini S., Riccio A., Santopolo S., et al. The FDA-approved drug nitazoxanide is a potent inhibitor of human seasonal coronaviruses acting at postentry level: effect on the viral spike glycoprotein. Front Microbiol. 2023;14 doi: 10.3389/fmicb.2023.1206951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vijgen L., Keyaerts E., Moës E., Maes P., Duson G., Van Ranst M. Development of one-step, real-time, quantitative reverse transcriptase PCR assays for absolute quantitation of human coronaviruses OC43 and 229E. J Clin Microbiol. 2005;43:5452–5456. doi: 10.1128/JCM.43.11.5452-5456.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Holzwarth G., Bhandari A., Tommervik L., Macosko J.C., Ornelles D.A., Lyles D.S. Vesicular stomatitis virus nucleocapsids diffuse through cytoplasm by hopping from trap to trap in random directions. Sci Rep. 2020;10 doi: 10.1038/s41598-020-66942-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Loo S.L., Wark P.A.B., Esneau C., Nichol K.S., Hsu A.C., Bartlett N.W. Human coronaviruses 229E and OC43 replicate and induce distinct antiviral responses in differentiated primary human bronchial epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2020;319:L926–L931. doi: 10.1152/ajplung.00374.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Karousis E.D., Schubert K., Ban N. Coronavirus takeover of host cell translation and intracellular antiviral response: a molecular perspective. EMBO J. 2024;43:151–167. doi: 10.1038/s44318-023-00019-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dolliver S.M., Galbraith C., Khaperskyy D.A. Human betacoronavirus OC43 interferes with the integrated stress response pathway in infected cells. Viruses. 2024;16:212. doi: 10.3390/v16020212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dolliver S.M., Kleer M., Bui-Marinos M.P., Ying S., Corcoran J.A., Khaperskyy D.A. Nsp1 proteins of human coronaviruses HCoV-OC43 and SARS-CoV2 inhibit stress granule formation. PLoS Pathog. 2022;18 doi: 10.1371/journal.ppat.1011041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Krähling V., Stein D.A., Spiegel M., Weber F., Mühlberger E. Severe acute respiratory syndrome coronavirus triggers apoptosis via protein kinase R but is resistant to its antiviral activity. J Virol. 2009;83:2298–2309. doi: 10.1128/JVI.01245-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yan X., Hao Q., Mu Y., et al. Nucleocapsid protein of SARS-CoV activates the expression of cyclooxygenase-2 by binding directly to regulatory elements for nuclear factor-kappa B and CCAAT/enhancer binding protein. Int J Biochem Cell Biol. 2006;38:1417–1428. doi: 10.1016/j.biocel.2006.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 61.Liu M., Yang Y., Gu C., et al. Spike protein of SARS-CoV stimulates cyclooxygenase-2 expression via both calcium-dependent and calcium-independent protein kinase C pathways. Faseb J. 2007;21:1586–1596. doi: 10.1096/fj.06-6589com. [DOI] [PubMed] [Google Scholar]
- 62.Chen J.S., Alfajaro M.M., Wei J., et al. bioRxiv; 2020. Cyclooxygenase-2 is induced by SARS-CoV-2 infection but does not affect viral entry or replication. [Preprint] [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.