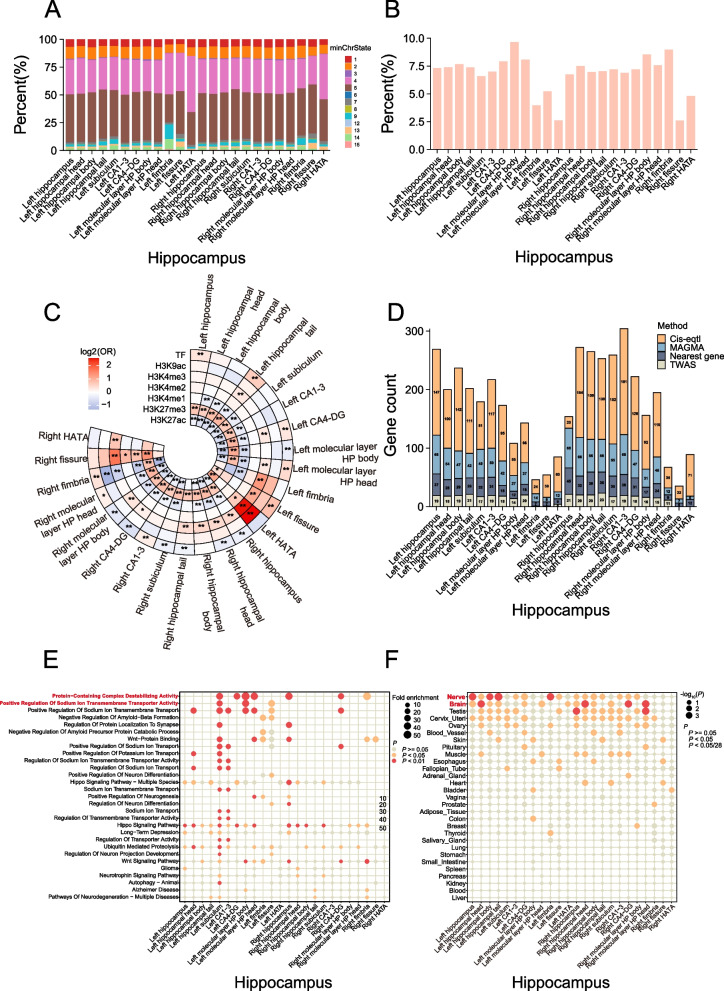
Fig. 3.
Annotation of risk variants and genes. A The minimum chromatin state across 127 tissue and cell types for candidate SNPs for each of the 24 HASV traits, with lower states indicating higher accessibility and states 1–7 referring to open chromatin states. B The bar charts represent the proportions of RegulomeDB scores 1 or 2 among the risk variants for each of the 24 HASV traits. A lower score suggests a higher likelihood of having a regulatory function. C Histone modifications and transcription factor (TF) peaks were primarily sourced from hippocampus samples provided by ENCODE. When hippocampus-specific data was unavailable, brain data was used instead. The enrichment of candidate SNPs in these epigenomic marks was assessed relative to control variants, which were generated in a 1:1 ratio using vSampler. Statistical significance was determined using a Fisher test. An asterisk denotes statistically significant differences (*P < 0.05; **P < 2.08 × 10−4, Bonferroni corrected). D The stacked bar charts depict the number of genes mapped using four distinct strategies: physical position, eQTL association, transcriptome-wide association study (TWAS), and multi-marker analysis of genomic annotation (MAGMA), for each of the 24 HASV traits. E Pathway analysis of genes associated with each of the 24 HASV traits based on the molecular signatures database. F Tissue expression results across 29 specific tissue types from GTEx v8 in FUMA. The chromatin states are 1 = active transcription start site (TSS); 2 = flanking active TSS; 3 = transcription at gene 5’ and 3’; 4 = strong transcription; 5 = weak transcription; 6 = genic enhancers; 7 = enhancers; 8 = zinc finger genes and repeats; 9 = heterochromatic; 10 = bivalent/poised TSS; 11 = flanking bivalent/poised TSS/Enh; 12 = bivalent enhancer; 13 = repressed PolyComb; 14 = weak repressed PolyComb; 15 = quiescent/low