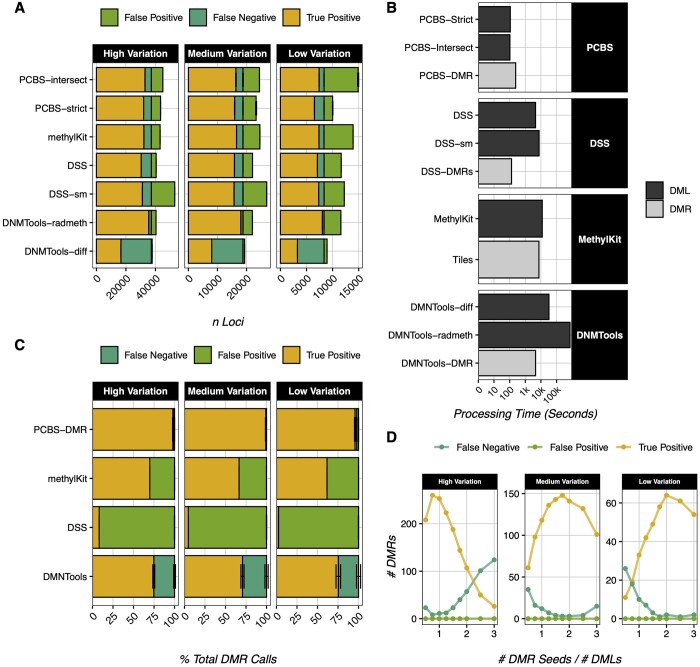
Figure 1.
PCBS accuracy and speed testing. (A) DML calling error rates of common WGBS software and PCBS in 127 iterations of 3 treatment versus 3 control samples for three simulated genomes of high, medium, and low variation; standard error of each call type is denoted with error bars. All DML calls use default software parameters. (B) Processing times of DML and DMR calling in common WGBS software and PCBS, on a single CPU with 8 GB RAM. All calls use default software parameters, and are run on eight archived mouse samples from Cole et al. (2017) aligned to mm10 at ∼30X coverage. (C) DMR calling error rates of common WGBS software and PCBS in 127 iterations of 3 treatment versus 3 control samples for three simulated genomes of high, medium, and low variation; standard error of each call type is denoted with error bars. DMR calls are determined to be true positives if they overlap any true DMR region. All DMR calls use default software parameters. (D) PCBS DMR calling accuracy under default parameters as a function of input seed number in one iteration of three treatment versus three control samples for three simulated genomes of high, medium, and low variation.