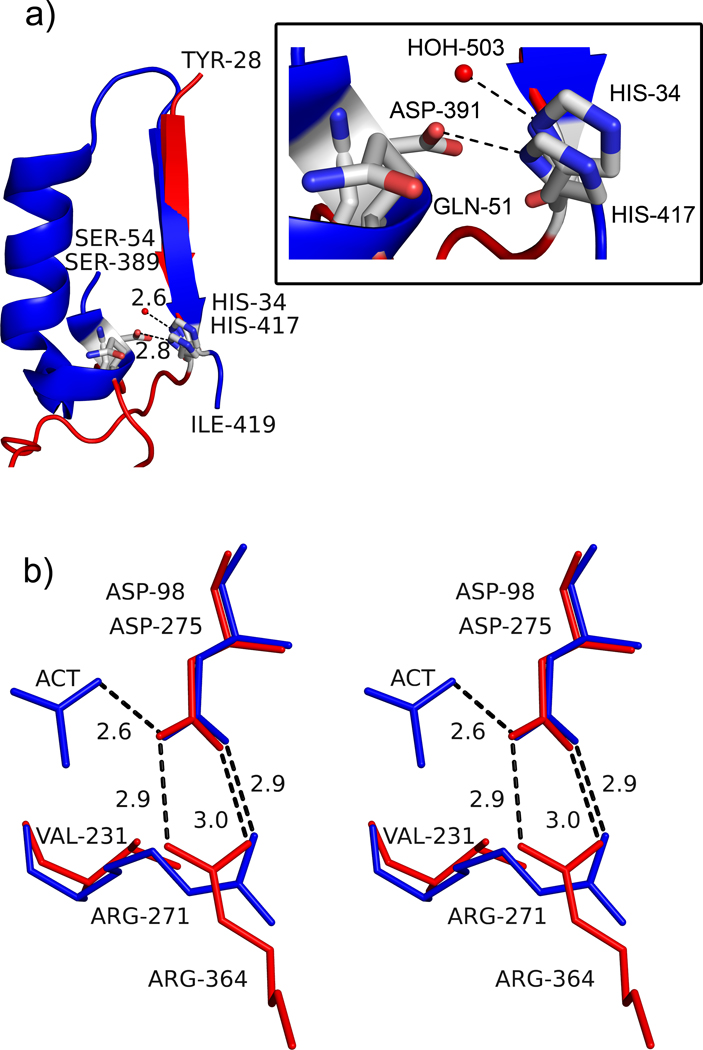
Figure 4.
Active site architecture in AaPgaBN. a) Part of the structure of AaPgaBN and SpPgdA showing MT5 motif and how the residues His34/His417 are presented into the active site. The red sphere represents the activating water molecule in the structure of AaPgaBN. The SpPgdA structure has a longer, more elaborate ordered region to project H417 and D391 whereas the AaPgaBN has a shorter less ordered segment to project His34 and Gln51 into the active site. The inset shows details of the interaction with residues and water molecule; b) Stereoview of the orientation and interaction of Arg271 to the catalytic base, Asp98 in AaPgaBN. Note the two electrostatic bonds between Arg271 and Asp98 (catalytic base shown in red) whereas the equivalent residues of SpPgdA, Arg364 and Asp275 (shown in blue), only have one. The direction of the interaction in AaPgaBN is altered and a Val231 is located at the position where Arg364 (SpPgdA) is present.