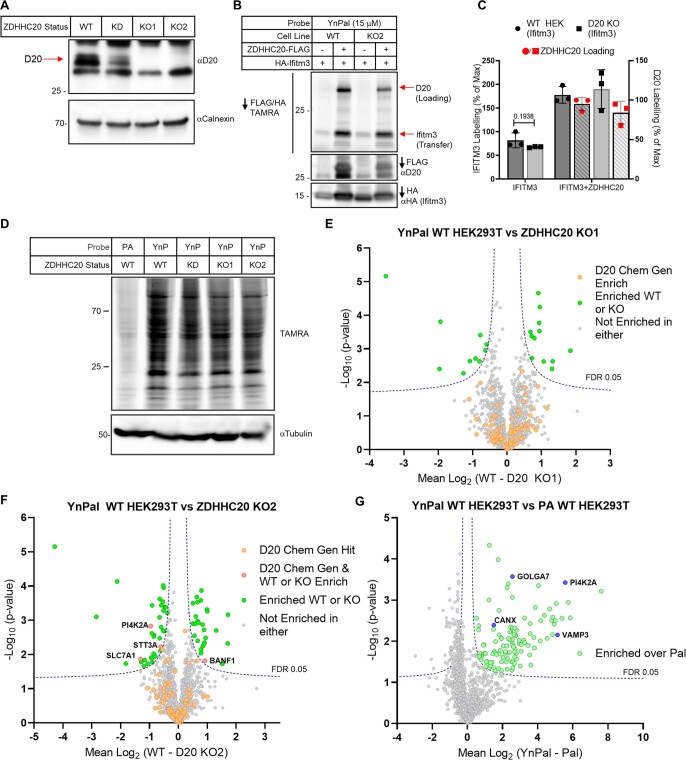
Extended Data Fig. 7. IFITM3 labeling in ZDHHC20 knock-out HEK293T cells.
(a) Untreated (UT) or gRNA/CAS9 treated (pSpCas9(BB)-2A-Puro, PX459 plasmid) HEK293T cells were probed with anti-ZDHHC20 (D20) and –vinculin antibodies. Cells treated with gRNA1/CAS9 resulted in knockdown (KD); whereas cells treated with gRNA2/CAS9 yielded two ZDHHC20-knockout (D20-KO) clones: KO1 and KO2 (n = 2 independent biological replicates). (b) WT or KO2 HEK293T cells were transfected with HA-IFITM3 and empty vector or C-FLAG-tagged ZDHHC20. Cells were then treated with 15 mM YnPal for 4 h before being harvested and lysed. IFITM3 and D20 were enriched in one pot with a mix of anti-HA and –FLAG resins before being treated with TAMRA-azide and click reagents. Tagged proteins were eluted from beads with 1X Laemmli buffer and separated by SDS-PAGE. YnPal ZDHHC20-loading and transfer to IFITM3 and input were visualized by in-gel fluorescence and anti-HA and -FLAG immunoblot, respectively (n = 2 independent biological replicates). (c) The average (n = 3 independent biological replicates) loading and transfer activity was reported as a percent of the maximal D20 fluorescent: input ratio and as a percent of the WT IFITM3 (empty vector) fluorescent: input ratio ± S.D. The two tailed unpaired t-test of Prism 9.0 was used to determine p-values and noted above relevant comparisons (d-g) WT HEK293T cells, two ZDHHC20 KO clones, and one partial knockdown (KD) clone were treated with 15 μM YnPal for 8 h. As a control for lipidation, HEK293T cells were treated with palmitic acid (Pal) and also taken through the experiment. Samples were then clicked with biotin-TAMRA-azide, 10% of which was analyzed by SDS-PAGE, in-gel fluorescence, and anti-tubulin western blot (d) (n = 3 independent biological replicates). The remainder was enriched on dimethylated neutravidin beads and digested for LC-MS/MS LFQ analysis. (E-G) Whilst a small number of proteins are identified as being significantly enriched/depleted (Student’s two tailed unpaired T-test S0 – 0.1, adjusted FDR – 0.05), they are few in number and none are consistently found which correspond to our putative chemical genetic substrates found in HEK293T cells. (f) Analysis of YnPal treated cells against Pal shows a large number a potentially lipidated proteins have been identified, with many well validated S-acylation proteins identified, some of which have been highlighted in blue.