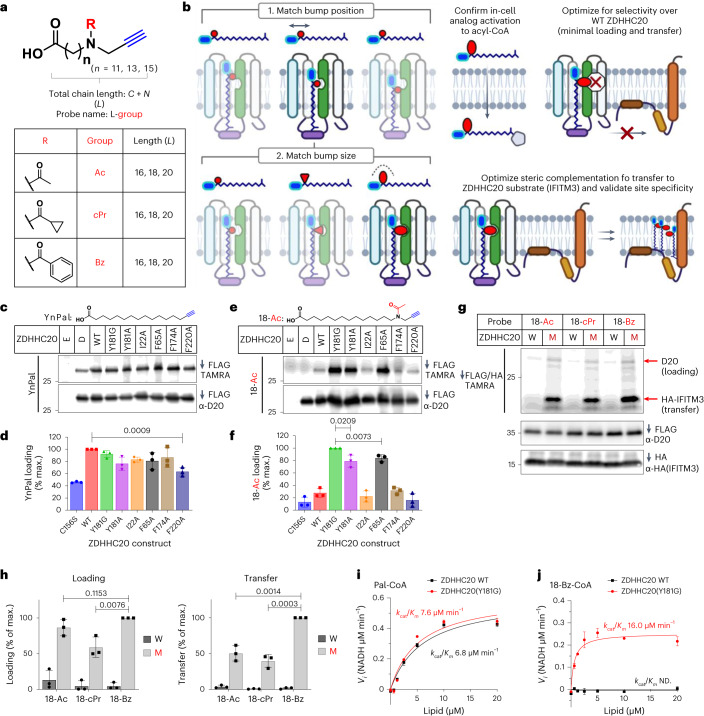
Fig. 2. Engineering a ‘bump’ probe and ‘hole’ mutant pair for ZDHHC20.
a, Fatty acid probes containing an alkynyl click-handle (blue), varying chain length L = 16, 18 or 20 heavy atoms in the chain (carbons + nitrogen) and an R ‘bump’ group (red)—Ac, cPr or Bz. b, Two-stage pairing strategy for a designed ZDHHC20 mutant optimizes probe chain length and then bump size to match the new binding cavity, with probe activation, selectivity over ZDHHC20 WT and transfer to a known ZDHHC20 substrate (IFITM3) optimized in parallel. c–f, Bump-hole loading analysis of C-terminal FLAG-tagged ZDHHC20 WT and mutants in HEK293T cells treated with 15 μM YnPal (c,d) or 18-Ac (e,f) for 4 h (D, catalytic-dead ZDHHC20(C156S); E, empty vector; n = 3 independent biological replicates average ± s.d.). g, Probe bump-size optimization by transfer assays with HA-IFITM3 and either WT ZDHHC20 (W) or ZDHHC20(Y181G) (M) co-expression in HEK293T cells (n = 3 independent biological replicates average ± s.d.). h, Average loading and transfer activity relative to highest fluorescent/input ratio (n = 3 independent biological replicates average ± s.d.). i,j, Enzyme kinetics for WT ZDHHC20 and ZDHHC20(Y181G) treated with Pal-CoA (i) or 18-Bz-CoA (j) using a KDH assay (3). Michaelis–Menten plots generated from average reaction rate (NADH generated μM min−1, n = 3 independent experiments) ± s.d. versus lipid concentration (μM). d,f,h, The two-tailed unpaired t test of Prism 9.0 was used to determine P values and noted above relevant comparisons.