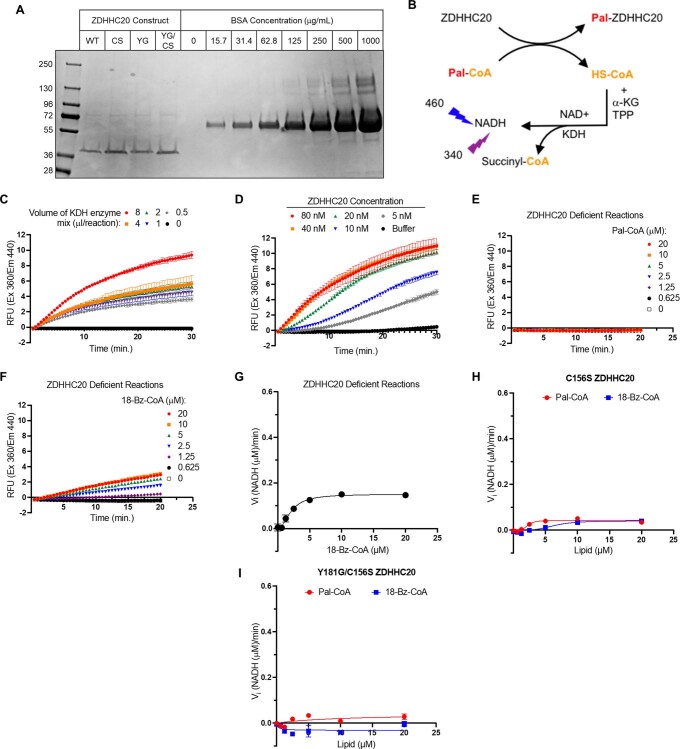
Extended Data Fig. 2. Establishing kinetic parameters for an optimal Y181G-ZDHHC20 bump probe pair.
(a) Wild-type (WT), Y181G (YG), C156S (CS) and Y181G/C156S (YGCS) FLAG-tagged ZDHHC20 constructs were transfected into HEK293T cells and purified by anti-FLAG agarose affinity chromatography. After enzyme elution with 3X FLAG-peptide, buffer was exchanged using 50 kDa M.W. cut-off protein concentrator tubes and sample concentration determined using a BSA standard curve. All samples were run on SDS-PAGE gels and protein visualized by Coomassie staining (n = 2 independent experiments). (b) An enzyme-coupled assay monitoring ZDHHC20 autoacylation was established using commercial α-ketoglutarate dehydrogenase enzyme (KDH) along with its substrates α-ketoglutarate (α-KG), thiamine pyrophosphate (TPP) and NAD+. Optimization of α-ketoglutarate dehydrogenase (KDH) (c) and WT ZDHHC20 (d) concentrations. Pal-CoA (e) and 18-Bz-CoA (f) KDH activities were determined in the absence of ZDHHC20, to establish background rates for each probe. (g) 18-Bz-CoA displayed significant background activity in the KDH assay without ZDHHC20. Reaction rates for ZDHHC20[C156S] (h) and ZDHHC20[Y181G, C156S] (i) treated with Pal-CoA or 18-Bz-CoA. Michaelis-Menten plots generated by plotting average (n = 3 independent experiments) reaction rates (NADH generated (µM)/min) ± S.D.) versus lipid concentration (µM) using Prism 9.0. For reactions with 18-Bz-CoA, the basal rates at all concentrations tested were subtracted from the corresponding total reaction rates.