Abstract
The assessment of the allergenic potential of chemicals, crucial for ensuring public health safety, faces challenges in accuracy and raises ethical concerns due to reliance on animal testing. This paper presents a novel bioinformatic protocol designed to address the critical challenge of predicting immune responses to chemical sensitizers without the use of animal testing. The core innovation lies in the integration of advanced bioinformatics tools, including the Universal Immune System Simulator (UISS), which models detailed immune system dynamics. By leveraging data from structural predictions and docking simulations, our approach provides a more accurate and ethical method for chemical safety evaluations, especially in distinguishing between skin and respiratory sensitizers. Our approach integrates a comprehensive eight-step process, beginning with the meticulous collection of chemical and protein data from databases like PubChem and the Protein Data Bank. Following data acquisition, structural predictions are performed using cutting-edge tools such as AlphaFold to model proteins whose structures have not been previously elucidated. This structural information is then utilized in subsequent docking simulations, leveraging both ligand–protein and protein–protein interactions to predict how chemical compounds may trigger immune responses. The core novelty of our method lies in the application of UISS—an advanced agent-based modelling system that simulates detailed immune system dynamics. By inputting the results from earlier stages, including docking scores and potential epitope identifications, UISS meticulously forecasts the type and severity of immune responses, distinguishing between Th1-mediated skin and Th2-mediated respiratory allergic reactions. This ability to predict distinct immune pathways is a crucial advance over current methods, which often cannot differentiate between the sensitization mechanisms. To validate the accuracy and robustness of our approach, we applied the protocol to well-known sensitizers: 2,4-dinitrochlorobenzene for skin allergies and trimellitic anhydride for respiratory allergies. The results clearly demonstrate the protocol’s ability to differentiate between these distinct immune responses, underscoring its potential for replacing traditional animal-based testing methods. The results not only support the potential of our method to replace animal testing in chemical safety assessments but also highlight its role in enhancing the understanding of chemical-induced immune reactions. Through this innovative integration of computational biology and immunological modelling, our protocol offers a transformative approach to toxicological evaluations, increasing the reliability of safety assessments.
Keywords: bioinformatics, agent-based models, predictive modelling, skin allergens, respiratory allergens, nonanimal methods, allergic asthma, regulatory toxicology
Introduction
Chemical allergies, resulting from exposure to low-molecular-weight chemicals, manifest as skin or respiratory reactions, posing significant health challenges. Traditionally, the identification of potential sensitizers has relied heavily on animal testing, which, besides ethical concerns, often fails to accurately predict human responses. This discrepancy underscores an urgent need for innovative methodologies in toxicological assessments.
Allergic contact dermatitis (ACD) is a common type of skin allergy resulting from chemical exposure, causing symptoms like redness, swelling, and blisters, affecting 15%–20% of the population [1]. Key allergens include metals, preservatives, and fragrances, with nickel, methylisothiazolinone, and thiurams, being notable examples [2].
ACD represents a delayed-type hypersensitivity reaction, primarily involving CD8+ Tc1/Tc17 and CD4+ Th1/Th17 effector T cells [3]. Extensively studied chemicals like 2,4-dinitrochlorobenzene (DNCB) have been instrumental in research, serving as reference skin sensitizers in cell-mediated immune reactivity studies since 1935 [4–6].
In contrast, respiratory allergies to low molecular weight (LMW) chemicals, though less prevalent, pose higher morbidity risks. Fewer than 100 chemicals are recognized as triggers, including diisocyanates and platinum salts [7, 8]. Respiratory sensitization lacks well-validated in vivo and nonanimal testing methods, making detection challenging [9].
Advances in understanding skin sensitization have led to the development and regulatory adoption of various testing methods. These include in vivo tests like the local lymph node assay [10] and in vitro methods such as the direct peptide reactivity assay (DPRA) [11–13]. These tests are part of a suite of approaches aiming to minimize animal testing and include regulatory frameworks [8–12].
Both types of sensitizers, despite causing distinct immune responses, share similarities. They are typically LMW chemicals that, being electrophilic or metabolizable to electrophilic species, react with proteins to initiate an immune response [14]. Most respiratory allergens have also shown reactivity in the DPRA, indicating overlapping detection methods for both allergy types [15, 16].
Research has established that skin sensitizers primarily induce Th1-type immune responses, whereas respiratory sensitizers are linked with Th2-type responses, evident from cytokine profiles [17–20]. This differentiation is supported by studies that evaluate cytokine profiles in the draining lymph nodes, using cytokines as biomarkers for identifying the type of sensitization [16, 21–26].
All these techniques mark progress by reducing animal use and focusing on molecular initiating events, like the covalent modification of proteins by allergens. However, they still do not capture the full complexity of the immune system’s response to allergens, particularly the interactions among various cell types and the subsequent cascade of immune signals [27].
To address these challenges, our research introduces a transformative computational approach that significantly enhances the field of toxicology. Central to our methodology is the integration of a sophisticated multistep bioinformatic protocol with the UISS. This innovative integration enables detailed simulations of the immunological impact of chemical exposures without relying on animal models.
UISS employs an agent-based modeling approach to simulate the intricate cellular and molecular dynamics of the human immune system in response to chemical sensitizers. This approach allows us to capture the specific pathways of immune activation, including the differentiation of T-helper cells (Th1, Th2, and Th17) and the secretion of cytokines that characterize either skin or respiratory sensitization. The ability to model these processes in silico offers a significant advantage over traditional in vivo methods, which often fail to capture such detailed immune responses. By mimicking the real-life behaviours and interactions of immune cells, UISS offers a dynamic representation of the immune response to chemical sensitizers, from initial exposure through the development of sensitization. A pivotal capability of UISS in our study is its ability to distinguish between Th1- and Th2-mediated immune responses—Th1 responses are more associated with cell-mediated immunity, crucial for skin sensitization scenarios, while Th2 responses are more fundamental in humoral immunity observed in respiratory allergies.
Moreover, UISS seamlessly integrates with upstream bioinformatic analyses, such as molecular docking and structural predictions [17]. This ensures that the simulator accounts for specific molecular interactions at the chemical–protein interface, using data on binding affinities [18, 19] and structural impacts to forecast the subsequent immune pathways. This integration enhances the predictive accuracy of chemical sensitization assessments, supporting the development of safer chemicals and aligning with global regulatory and ethical shifts towards reducing and replacing animal testing in chemical safety evaluations.
Incorporating UISS into our research protocol represents a major advancement in our ability to predict the immunological effects of chemical exposures accurately and ethically. This approach not only fills a critical gap in existing toxicological research but also sets a new standard in predictive accuracy and ethical conduct in chemical safety assessments.
By bridging detailed biochemical data with sophisticated immune system simulations, our approach promises to transform the landscape of chemical safety evaluation, enhancing both the scientific rigour and ethical standards of toxicological assessments.
Methods and materials
Workflow of the multistep and multiscale bioinformatic approach
The problem-solving protocol consists of three different phases that include eight distinct steps.
The first phase deals with data collection. Chemical structures of interest are sourced from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/). In parallel, selected tissue proteins that are relevant to the sensitization process are identified using the Human Protein Atlas (https://www.proteinatlas.org). For proteins whose structures are already known, databases such as the Protein Data Bank (PDB) (https://www.rcsb.org/) are consulted. If the structures are not known, AlphaFold (https://alphafold.ebi.ac.uk/) is employed to predict their 3D structures.
The second phase deals with the preprocessing and processing stages. Within this phase, the Chimera Software is used for preparing the docked configurations of the retrieved proteins. This preparation is critical for ensuring that the proteins are in the correct conformation for subsequent steps in the protocol. Next, the prepared proteins undergo a series of docking simulations. The simulations involve both ligands and proteins. The success of these interactions is quantified by docking scores, which serve as a metric to select the most promising ligand–protein complexes. This selection is not the end, however. The chosen complexes are then subjected to protein–protein docking procedures with a range of Toll-like receptors (TLRs). The outcome of these interactions is carefully recorded, highlighting complexes that exhibit the most favourable docking scores. The next step involves leveraging the Immune Epitope Database, or IEDB (https://www.iedb.org/), to predict potential epitopes of the complexes of interests, adding an extra layer of insight into how the immune system might recognize these compounds.
With the completion of the processing phase, the protocol shifts to the application of an in silico trial phase (third phase). This phase is characterized by the execution of UISS, which predicts the Th1, Th2, and Th17 immune responses, immunoglobulins, and massive cytokines panel dynamics. These predictions are instrumental in discerning the potential immunological response that the chemical compound might elicit.
UISS is an advanced computational framework designed for simulating the immune response within a bidimensional or tridimensional anatomical compartment. This simulator encapsulates each biological entity—ranging from pathogens (including viruses, bacteria, fungi, and helminths), and malignant cellular transformations, to immune response cells—as autonomous agents. These agents are distinguished by specific characteristics such as type, admissible states, spatial positioning, and interaction patterns. The simulator employs stoichiometric equations to model potential chemical reactions among species, thereby facilitating a stochastic representation of physiological interactions within the immune system and disease mechanisms. The UISS operates on the principle of agent-based modelling (ABM), a simulation technique that allows for the individual tracking of entities, contrary to aggregate models like differential equations. This approach enables the emergence of complex behaviours from simple rules, offering insights into nonintuitive system dynamics. The simulation progresses over discrete time intervals, with the system’s evolution influenced by stochastically determined interactions, movement, and internal state changes of agents. This evolution adheres to statistical determinism, meaning that while individual simulation runs may vary due to randomness, the average of numerous runs converges to a predictable outcome, consistent with the central limit theorem. At the core of the UISS is a detailed representation of the immune system’s cellular and molecular constituents, with mechanisms such as haematopoiesis, thymus education (maturation and differentiation of T cells, both positive and negative selections are taken into account), antigen recognition, and immune memory faithfully modelled. The system’s spatial dynamics are captured through a lattice framework, where agents can move and interact within defined compartments like the thymus, bone marrow, and secondary organs. Interactions are governed by rules that consider proximity, chemical concentration gradients, and specific receptor–ligand affinities, with outcomes affecting the states and fates of involved entities. The simulator is developed in ANSI C-99, ensuring portability across different computer architectures. By leveraging ABM’s strengths, the UISS offers a powerful means to explore the immune system’s response to various pathogens and conditions, with the flexibility to incorporate new biological insights and complex interactions. This enables a more nuanced understanding of immune dynamics, potentially guiding the development of therapeutic strategies.
Moreover, UISS accounts for biological variability in immune responses. In particular, it incorporates genetic variability by allowing the customization of parameters that influence immune responses. These parameters can be adjusted to reflect differences in genetic makeup, such as variations in major histocompatibility complex (MHC) molecules, cytokine production levels, and receptor expression. The model can also simulate the impact of environmental factors, such as prior exposure to allergens, infections, and overall health status, which can significantly influence an individual’s immune response. These factors are incorporated through initial conditions and external stimuli inputs.
UISS-TOX, or the Universal Immune System Simulator for Toxicology, is an extension or application of the UISS framework specifically designed to study the effects of toxic substances on the immune system. While the original UISS model is focused on simulating the immune system’s response to pathogens, cancerous cells, and other internal biological entities, UISS-TOX adapts this sophisticated simulation environment to understand how toxins and potentially harmful chemicals interact with immune system components [20, 25]. By incorporating toxicological factors into the UISS model, UISS-TOX enables the simulation and the analysis of the impact of various toxic substances on immune function. This can include how toxins affect the behaviour of immune cells, alter chemical signalling pathways, or disrupt normal physiological responses. The simulation can cover a wide range of toxicological scenarios, from acute exposure to long-term effects, offering insights into how toxic substances might compromise the immune system’s ability to fight infections, contribute to the development of autoimmune diseases, or influence the progression of cancer.
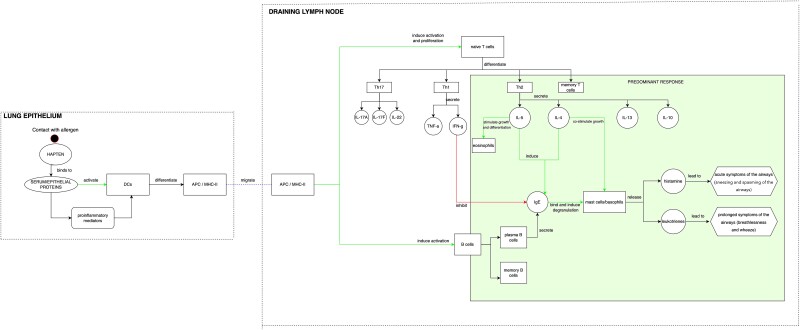
We have further extended the UISS-TOX modelling and simulation platform to consider relevant hallmarks of skin and respiratory sensitizers. To achieve this goal, we have systematically reviewed the literature to identify the primary immunological mechanisms behind allergic reactions and have illustrated these mechanisms within a conceptual framework (Fig. 1). The model depicts the key immunological events of an allergic reaction, with a particular focus on respiratory Th2-type reactions. The two main represented compartments are lung epithelium and draining lymph node. After exposure, the respiratory sensitizing chemical in lung epithelium binds to serum or epithelial proteins. The primary haptenized proteins for respiratory sensitizers were found to be multiple unknown serum proteins with molecular weights of 130 000 and above, as well as electrophoretic mobility and coelution with the prominent proteins in serum albumin [26]. This process, helped by proinflammatory mediators, activates dendritic cells (DCs), which differentiate in antigen-presenting cells/MHC-II. The antigen-presenting cells migrate to the draining lymph node, where they induce the activation of B cells and the proliferation of naive T cells, promoting their differentiation in Th1, Th2, and Th17. In the case of respiratory sensitization, the differentiation is more driven towards Th2 cells, with the secretion of cytokines such as IL-5, IL-4, IL-13, and IL-10. IL-5 stimulates the growth and differentiation of eosinophils and, together with IL-4, induces IgE secretion from plasma B cells. IgE binds and induces degranulation of mast cells and their release of histamine and leukotrienes. In particular, histamine leads to the acute symptoms of the airways (sneezing and spasms of the airways), while leukotrienes lead to the prolonged symptoms of the airways (breathlessness and wheezing), all characteristic symptoms of respiratory allergy.
Figure 1.
Conceptual model representation of the key immunological events of skin and respiratory sensitization, focusing on a Th2-type respiratory predominant response.
UISS-TOX accepts as an input a datafile containing, among other specific immune system parameters, the computed docking scores regarding chemicals under investigation and the predicted epitopes that potentially elicit immunogenicity.
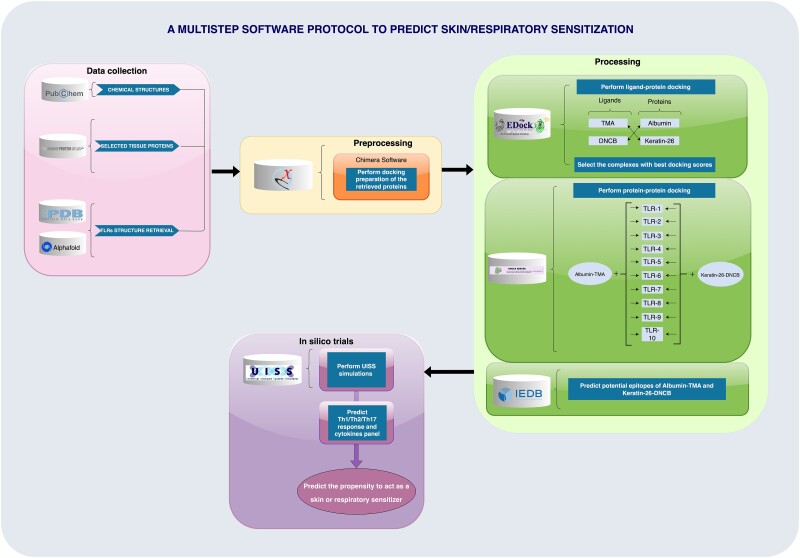
Figure 2 summarizes the protocol, highlighting the three phases (data collection, processing, and in silico trial).
Figure 2.
An integrated bioinformatic approach to predict skin and respiratory reactions. This multistep protocol leverages the synergy of computational tools to forecast the sensitizing potential of chemicals. Beginning with a robust data collection from PubChem and protein structures from PDB and AlphaFold, it advances through meticulous preprocessing with Chimera Software for docking. The processing core utilizes EpiDock to explore ligand–protein interactions, with subsequent protein–protein docking forming a complex web of TLR interactions. Finally, in silico trials run on UISS software not only predict the mounting of a specific helper T-cell and key cytokine responses but also determine the likelihood of a substance to act as a sensitizer, streamlining the identification process for safer product development.
Table 1 lists the software used in the workflow, along with their functions and purposes, and provides the corresponding online URLs or download links.
Table 1.
Software used in the workflow.
| Software | Purpose | Link |
|---|---|---|
| ChimeraX | Perform docking preparation of the retrieved proteins (albumin and Keratin-26). | https://www.cgl.ucsf.edu/chimerax/download.html |
| EDock | Perform blind protein–ligand docking between each chemical and each retrieved protein. | https://zhanggroup.org/EDock/ |
| HDock | Perform protein–protein blind docking between the complexes with the beast docking scores and each TLR. | http://hdock.phys.hust.edu.cn/ |
| IEDB | Predict potential epitopes from the complexes with the best docking scores. | https://www.iedb.org/ |
| UISS | Simulate Th1/Th2/Th17 response and cytokine panels. | https://combine-group.org/ |
The results generated from this comprehensive protocol do not merely predict outcomes but offer a profound understanding of the immunological mechanisms involved. These insights have substantial implications for further research and development, profoundly impacting product safety evaluations and supporting a shift towards more ethical, nonanimal-based methods in toxicology. In what follows, we apply the described protocol to a specific case study, with the aim to show how it performs and to provide a detailed user guide to practitioners and researchers who would like to use the methodology.
A working example: applying the protocol to predict the skin or respiratory allergic reactions to trimellitic anhydride and 2,4-dinitrochlorobenzene chemicals
In what follows, we describe a concrete application of the three-phase, eight-step protocol to predict and discriminate allergic reactions to two chemicals: DNCB and TMA.
Phase 1—data collection
Retrieve the chemical structure of the unknown potential sensitizer
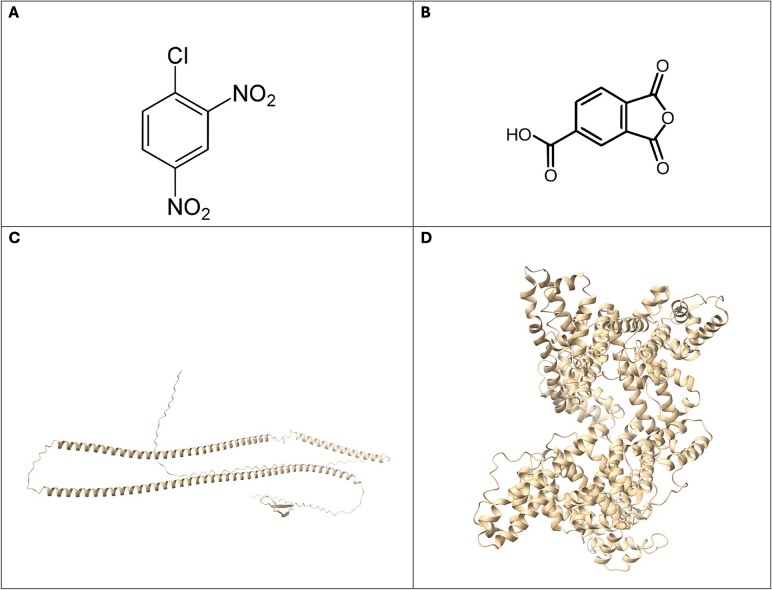
The initial step of the pipeline involves obtaining the chemical structure of the unknown potential sensitizer. In our case study, we selected DNCB and TMA because they are well-known skin and respiratory sensitizers, respectively. We retrieved the chemical structures of DNCB (Fig. 3A) and TMA (Fig. 3B) from PubChem, an open chemistry database maintained by the National Institutes of Health (https://pubchem.ncbi.nlm.nih.gov/).
Figure 3.
(A) and (B) represent, respectively, the DNCB and TMA chemical structures (source: PubChem). (C) and (D) display, respectively, Keratin-26 and human serum albumin 3D structures (source: PDB).
Retrieve potential proteins representative of different biological compartments
The second step consists of retrieving protein structures representative of different biological compartments. In our case study, we considered the human serum albumin (Fig. 3D), as it is the major immunologically relevant protein conjugated in exposed humans affected by respiratory sensitization [26], and Keratin–26 (Fig. 3C), as it is a highly expressed epithelial protein in the skin. We retrieved the 3D structure of human serum albumin from Protein Data Bank (https://www.rcsb.org/), which is a comprehensive online repository that stores 3D structural data of biological macromolecules, such as proteins and nucleic acids, derived from experimental methods like X-ray crystallography, nuclear magnetic resonance (NMR) spectroscopy, and cryo-electron microscopy. The Keratin-26 3D structure was retrieved from The Human Protein Atlas (https://www.proteinatlas.org/), a database of proteins consisting of 12 separate sections, each focusing on a particular aspect of the genome-wide analysis of the human proteins.
Retrieve the structures of the main Toll-like receptors
TLRs are integral components of the innate immune system, typically recognized for their role in detecting and responding to microbial pathogens. However, recent research has unveiled their involvement in allergic reactions, particularly in the skin and respiratory system [28]. In the skin, TLRs are expressed in cells such as keratinocytes and DCs. They play a role in maintaining the skin’s barrier function and can trigger immune responses upon detecting allergens or danger signals from them. In conditions like atopic dermatitis (eczema), chronically activated TLR signalling pathways may contribute to chronic inflammation [29].
Similarly, in the respiratory system, TLRs are expressed on epithelial cells lining the airways. They recognize both microbial components and environmental allergens. Activation of TLRs on these cells can lead to the production of inflammatory mediators, contributing to allergic inflammation seen in asthma and allergic rhinitis [30].
Moreover, TLRs are also present in various immune cells in the respiratory tract, such as DCs, macrophages, and mast cells [31, 32]. Activation of these receptors by allergens or other stimuli can modulate the immune response, further influencing the development and severity of respiratory allergies [28]. For these reasons, the structures of TLRs from AlphaFold were retrieved (https://alphafold.ebi.ac.uk/). AlphaFold is an artificial intelligence system developed by Google DeepMind, able to computationally predict protein structures with accuracy and speed [33]. For the sake of completeness, all TLRs involved in the human immune system were used, i.e. TLR1 to TLR10.
Phase 2—processing
Perform docking preparation of the retrieved proteins
Prior to docking, a crucial preparatory phase involves meticulously handling both the ligand and the receptor. This step entails processes such as removing ligand and solvent molecules, rectifying alternate locations of residues, converting selenomethionines to methionines, introducing hydrogen atoms, assigning charges to protein atoms, checking for incomplete residues, and substituting them with glycines. We used the ‘Dock Prep’ tool of Chimera X suite [34] for docking preparation, which performs several tasks to prepare structures for molecular docking or other calculations, including deleting water molecules, repairing truncated sidechains, adding hydrogens, and assigning partial charges.
Perform blind protein–ligand docking using EDock server
Following the completion of docking preparations, we employed the EDock server [35] to execute blind protein–ligand docking for the following pairs: (i) albumin-TMA, (ii) albumin-DNCB, (iii) keratin-26-TMA, and (iv) keratin-26-DNCB. In this process, albumin and keratin-26 were used as receptors, while DNCB and TMA served as ligands. EDock is a high-quality blind docking system based on low-resolution protein structure prediction, which uses replica-exchange Monte Carlo (REMC) simulations. Using a query protein sequence as a starting point, I-TASSER is then used to estimate the target protein’s 3D model, from which COACH can predict the ligand binding site. A modified graph matching is used to produce the initial ligand poses based on the predicted binding pockets. Under the direction of a physical force field combined with binding site constraints, REMC simulations are carried out for ligand conformation sampling; the ligand docking model is ultimately chosen using a composite knowledge-based score function.
The EDock server provided us with outputs including the predicted ligand binding sites by COACH and the docking poses by EDock (the final docking poses were generated by cluster 1 of the COACH output). We retrieved the various scores provided by EDock, and we selected the ligand–protein complexes having the highest docking score in terms of ‘X SCORE’, which is the scoring function utilized by EDock for ranking the docking poses.
Perform protein–protein blind docking using HDock server
The following step consisted in performing blind protein–protein docking. Specifically, we focused on the two complexes with the highest docking scores from the previous step, i.e. albumin-TMA and keratin-26-DNCB. We used HDock server [36] to dock these complexes with all the retrieved TLRs to explore potential interactions. HDock allows to perform protein–protein and protein–DNA/RNA docking based on a hybrid algorithm of template-based modelling and ab initio free docking. We used the PDB structures of the complexes albumin-TMA and keratin-26-DNCB as ligands, and the PDB structure of each TLR as receptor. We conducted docking simulations with each ligand paired with every TLR, and we collected the scores of each docking simulation.
Predict potential epitopes through Machine Learning/Artificial Intelligence approaches (Immune Epitope Database)
Once protein–protein blind docking scores have been obtained, we performed structure-based prediction of antibody epitopes of the Albumin-TMA and Keratin-26-DNCB poses, using ElliPro [37]. ElliPro aims to predict and visualize antibody, or B-cell, epitopes within a given protein sequence or structure. ElliPro integrates Thornton’s method with a residue clustering algorithm, the MODELLER program for structural prediction, and the Jmol viewer for the visualization of predicted epitopes. The tool’s ability to consistently rank its best prediction within the top three for a significant majority of proteins underlines its utility in the identification of antibody epitopes for the Albumin-TMA and Keratin-26-DNCB proteins antigens.
Phase 3—in silico trial
Predict Th1/Th2/Th17, immunoglobulins, and cytokine panel dynamics
After steps 1–7 are completed, UISS-TOX is now ready to predict Th1, Th2, Th17, and immunoglobulins immune responses alongside a comprehensive panel of cytokines. This advanced capability is instrumental in distinguishing and accurately predicting whether a chemical under investigation is more likely to act as a contact sensitizer or respiratory sensitizer, by analyzing the predominant T-helper cell polarization and cytokine profiles. While skin sensitization can involve both Th1 and Th2 responses depending on the specific condition, such as the Th2 dominance in atopic dermatitis and the Th1 dominance in ACD, respiratory sensitization is typically associated with a predominant Th2 polarization. Through this comprehensive assessment, UISS-TOX is poised to deliver nuanced insights into the immune response elicited by chemicals, thereby offering a ground-breaking approach to discerning their sensitizing potential.
All these eight steps are correlated and connected. The chemical structure retrieval directly informs the docking preparation and simulations by providing the initial data needed for interaction predictions.
The docking preparation ensures that proteins are in a suitable state for accurate protein–ligand and protein–protein docking, which are critical for predicting how chemicals interact with biological molecules.
The docking scores and epitope predictions provide specific data inputs for the UISS-TOX simulations, ensuring that the immune response predictions are based on detailed molecular interactions.
Results and discussion
This study delineates a computational workflow designed to discriminate between skin and respiratory sensitizers by leveraging their distinct T-helper responses—Th1 for skin and Th2 for respiratory. Existing methods lack the capability to make such distinctions; thus, our approach uses DNCB, a known skin sensitizer, and TMA, recognized for its respiratory sensitization properties, to validate the effectiveness and precision of our model.
The workflow consists of three phases. The first phase involves a comprehensive data collection, extracting chemical structures of DNCB and TMA from the PubChem database and relevant protein structures from Protein Data Bank and The Human Protein Atlas. This includes human serum albumin, crucial for respiratory sensitization, and Keratin-26, a key epithelial protein. Postdata retrieval, the second phase includes preprocessing and processing stages, involving software like Chimera X for docking preparations, essential for the subsequent accuracy of docking simulations, and servers such as EDock and HDock for docking simulations. This step is crucial for understanding the molecular basis of binding affinities, which can significantly influence the efficacy of potential therapeutic compounds. Docking servers such as EDock and HDock further streamline the simulation process, as demonstrated in recent studies. For instance, Paul et al. [18] utilized these platforms to investigate the anthelmintic activity of pineapple-derived compounds. Through a series of molecular docking experiments and molecular dynamics simulations, they evaluated the stability and binding interactions of ligands with target proteins, validating the inhibitory potential of key compounds against parasitic enzymes. Similarly, Obaidullah et al. [17] performed molecular docking to explore the interaction of bioactive phytochemicals from Cnesmone javanica with anxiolytic and antidepressant receptors, underscoring the importance of these in silico tools for drug discovery. In addition to molecular docking, density functional theory (DFT) calculations have become integral to computational drug discovery. These methods provide valuable insights into the electronic properties of molecules. For instance, Rahman et al. [19] applied DFT calculations to study the antidepressant activity of compounds from Cycas pectinata, revealing the importance of frontier molecular orbitals and vibrational frequencies in understanding compound reactivity.
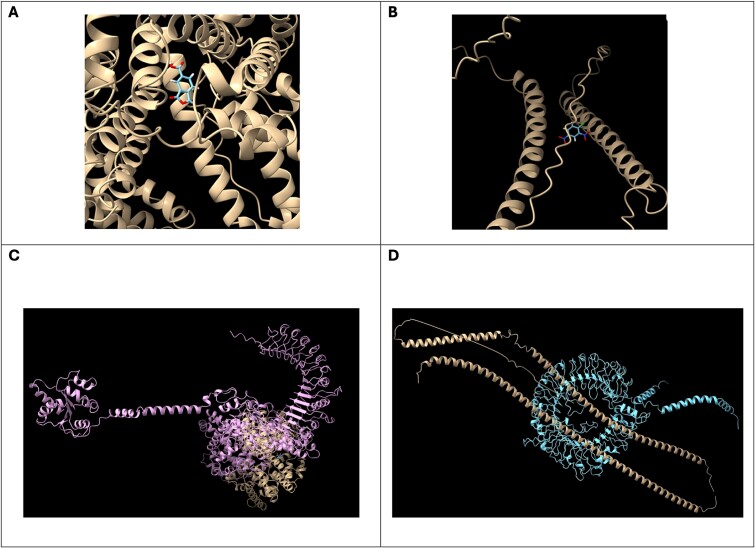
EDock server is used to assess interactions between each ligand and protein, resulting in notable complexes: albumin-TMA and keratin-26-DNCB (Fig. 4A and B, respectively). Following this, the HDock server facilitates protein–protein docking between these complexes and a range of TLRs (TLR1–TLR10), which play significant roles in immune responses to allergens in both skin and respiratory systems (Fig. 4C and D, respectively).
Figure 4.
(A) Best docked pose of albumin-TMA predicted by EDock; (B) best docked pose of Keratin-26-DNCB predicted by EDock; (C) albumin-TMA + TLR2 structure interaction predicted by HDock; and (D) Keratin-26-DNCB + TLR9 structure interaction predicted by HDock.
Further analysis involves using ElliPro for structure-based prediction of antibody epitopes of the albumin-TMA and Keratin-26-DNCB complexes (Fig. 5A and B, respectively). The docking scores and predicted epitopes are subsequently used as inputs for UISS-TOX, enhancing our understanding and prediction of chemical sensitization pathways, critical for advancing safety assessments in toxicology.
Figure 5.
(A) Best scored predicted linear epitopes for albumin-TMA structure and (B) best scored predicted linear epitopes for Keratin-26-DNCB structure.
To assess the sensitizing potential of skin or respiratory sensitizers, we employed UISS-TOX modelling and simulation infrastructure as an in silico trial to inform whether a specific chemical can drive a skin or respiratory sensitizer pathway.
According to the obtained docking scores, we report for both Albumin-TMA and Keratin-26-DNCB with TLRs 1–10 the Th1, Th2, and Th17 immune responses alongside a comprehensive panel of cytokines.
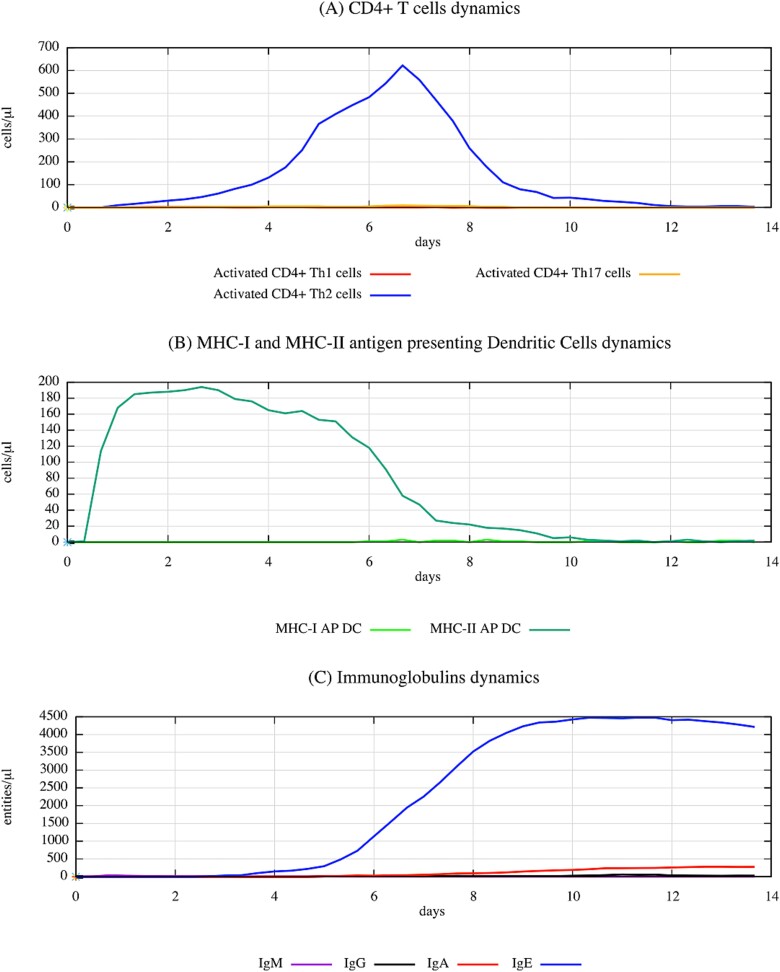
The dynamic interplay in the immune response to the Albumin-TMA complex is depicted in Fig. 6, showcasing robust Th2 and IgE responses over a 14-day period. The rise in Th2 cells and IgE levels indicates a strong allergic reaction, consistent with what one can expect from a respiratory sensitizer like TMA. Interestingly, while Th2 responses dominate, there are measurable responses in Th1 and Th17 subpopulations, along with various immunoglobulin classes, especially IgE, showing the complexity of the immune response.
Figure 6.
Dynamic interplay in immune response to albumin–TMA complex as simulated by UISS-TOX, highlighting the robust TH2 and IgE responses over a 14-day period. This figure illustrates the evolution of DCs, CD4 T-cell subpopulations (TH1, TH2, and TH17), and various immunoglobulin titers. Notably, TH2 cells and IgE titers show prominent increases, indicative of a strong allergic-type immune reaction typical of respiratory sensitizers. Concurrently, other immune responses, including TH1 and TH17 subpopulations, along with IgM, IgG, and IgA subclasses, are also present but to a lesser degree, emphasizing the nuanced and multifaceted nature of immunological responses. The simulation’s prediction aligns with the characterization of TMA as a respiratory sensitizer, underscoring the significant role of TH2 responses in this determination.
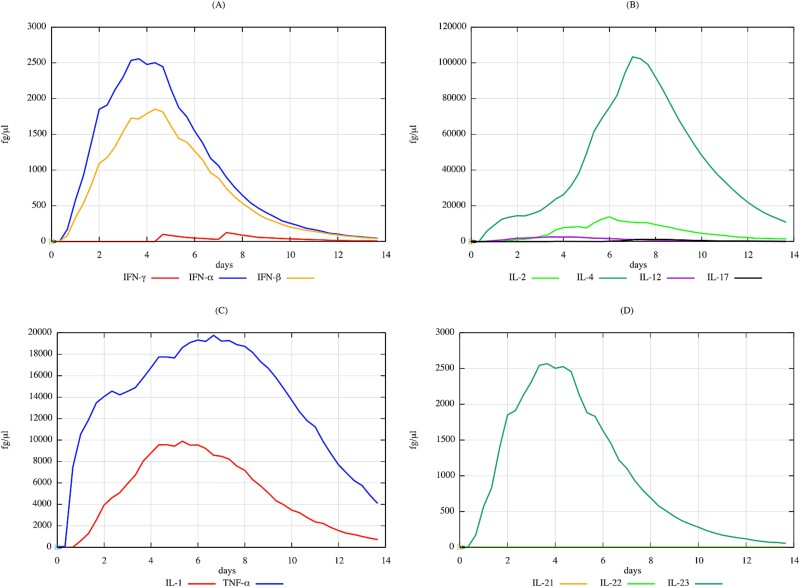
The cytokine levels in response to the same albumin–TMA complex can also provide valuable insights. The significant increase in IL-4 and IL-17, as depicted in Fig. 7, suggests a robust Th2-mediated response and potential contribution to inflammatory processes. The cytokine profile underlines the intricate network of immune signalling that takes place in reaction to respiratory sensitizers, with Th2 responses being significant but not exclusive.
Figure 7.
Comprehensive prediction of cytokine dynamics in response to albumin–TMA complex over a 14-day period as simulated by UISS-TOX3. This figure details the fluctuations in key cytokines, including IFN-gamma, IFN-alpha, IFN-beta, IL-2, IL-4, IL-12, IL-17, IL-1, TNF, IL-21, IL-22, and IL-23. Notably, there are significant elevations in IL-4 and IL-17, suggesting both strong TH2-mediated responses and potential involvement in inflammatory processes. The presence of varied cytokine responses underscores the complex interplay of immune signalling pathways, with implications for understanding the inflammatory and allergic responses to respiratory sensitizers like TMA.
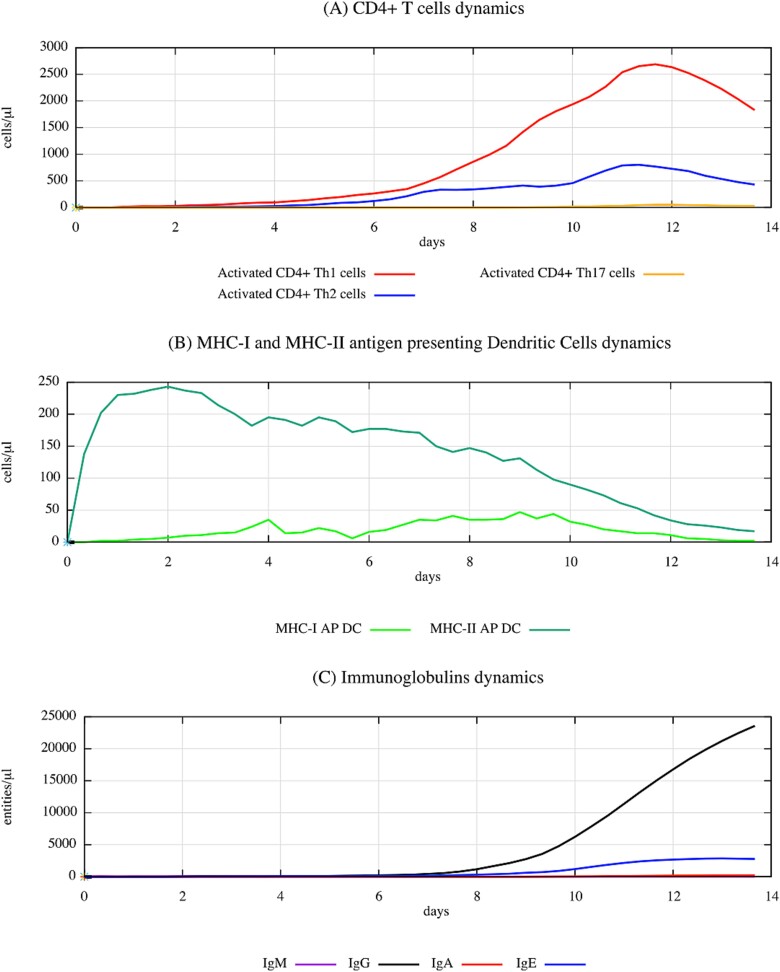
Concerning the keratin–DNCB complex, the related in silico predictions provide insights about the cellular and humoral components involved in ACD, as one can observe in Fig. 8. The significant activation of Th1 cells supports the profile of ACD as a Th1-mediated response. This is a severe contrast to the Th2 dominance observed in Fig. 6, indicative of different immune pathways activated by different sensitizers.
Figure 8.
Simulated immune dynamics in response to keratin-DNCB complex over a 14-day period, highlighting the cellular and humoral activities characteristic of ACD sensitization, as modelled by UISS-TOX. This figure captures the progression of DCs, various CD4+ T-cell subsets (TH1, TH2, TH17), and immunoglobulin responses (IgM, IgG, IgA, IgE). The data show a significant activation of TH1 cells, consistent with the TH1-mediated immune reaction typical in ACD.
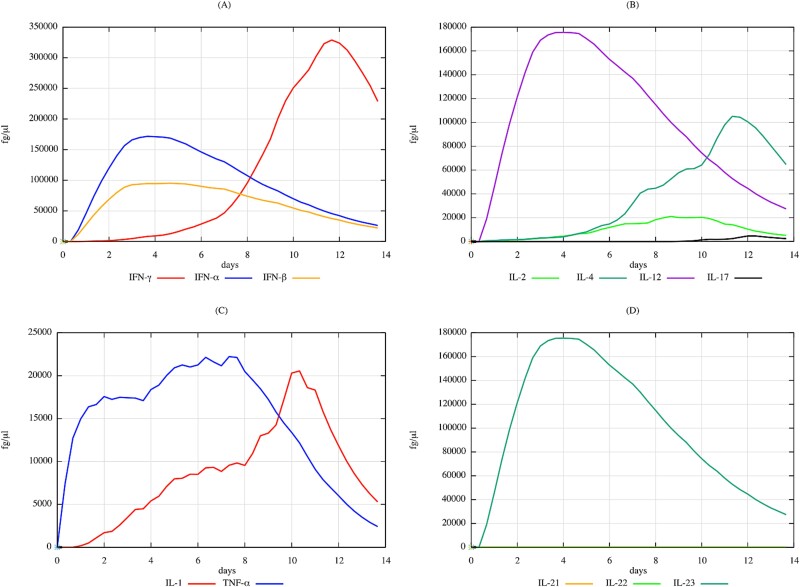
In parallel, the cytokine profile for the Keratin–DNCB complex over 14 days indicates a pronounced inflammatory reaction, with significant elevations in pro-inflammatory cytokines such as interferon (IFN)-gamma, tumor necrosis factor (TNF), and a range of interleukins (Fig. 9). These findings are consistent with the key drivers of the ACD response and reflect the critical role of these cytokines in the sensitization process associated with skin exposure.
Figure 9.
Quantitative simulation of cytokine profiles over 14 days in response to keratin–DNCB complex, elucidating the immunological pathways involved in ACD as modelled by UISS-TOX3. This figure reveals significant elevations in cytokines such as IFN-gamma, TNF, and various interleukins (IL-1, IL-2, IL-12, IL-17, IL-21, IL-22, IL-23), reflecting the complex inflammatory network activated by DNCB. The heightened levels of pro-inflammatory cytokines, particularly TNF and IL-17, highlight the key drivers of the ACD response, emphasizing the critical role of these cytokines in skin sensitization processes.
In summary, the UISS-TOX and related extended modules offer a comprehensive prediction of immune responses, from cellular activities to cytokine profiles, elucidating how various substances may trigger allergic or sensitization responses. These models serve as pivotal components of our proposed advanced computational protocol, designed to discriminate whether an unknown chemical has the potential to act as a skin or respiratory sensitizer. Our preliminary results with DNCB, a well-documented skin sensitizer, and TMA, a known respiratory sensitizer, demonstrate that the models can accurately distinguish between these sensitization pathways. The outcomes are not only valuable for predicting potential health risks associated with chemical exposure, thereby reducing the need for in vivo testing, but also provide quantitative information crucial for risk assessment. As further validation with additional chemicals continues, the UISS models are poised to be integral to an Integrated Approach to Testing and Assessment, identifying chemicals that exert toxic effects through immunological mechanisms and determining the likely type of immune response elicited.
Conclusions
The integration of in silico methods into toxicological assessments represents a substantial leap forward in predictive toxicology. This study has demonstrated that our computational pipeline fills a critical gap in accurately predicting chemical-induced allergic reactions. By providing a reliable and efficient alternative to traditional animal-based testing, our protocol not only accelerates the testing process but also significantly reduces the ethical, environmental, and financial burdens associated with conventional toxicology.
Our findings confirm that the UISS model can accurately predict immune responses to chemical sensitizers, effectively distinguishing between Th1-driven skin reactions and Th2-driven respiratory sensitizations. This ability to differentiate between immune pathways is a major advancement over existing methods, which often lack this level of precision. The successful validation of our model with well-established sensitizers, DNCB and TMA, underscores its robustness and potential for widespread application. Importantly, this approach offers a viable and ethical alternative to animal testing, directly addressing the growing need for nonanimal methods in regulatory toxicology. By integrating detailed molecular docking data with advanced immune system simulations, the approach captures the complex interactions and signalling pathways involved in allergic reactions, offering a deeper understanding of the underlying mechanisms. The ability to simulate the entire immune response, including cytokine dynamics and T-cell differentiation, enhances the understanding of how chemical exposures can lead to sensitization, providing valuable insights for developing safer chemicals and therapies.
The advantages of this approach are significant and multifaceted. By aligning with global regulatory and ethical initiatives to reduce and replace animal testing, our computational model offers a humane and scientifically robust alternative for chemical safety assessment. Furthermore, in comparison to traditional in vivo methods, computational modelling is not only faster and more cost-effective but also provides greater flexibility in testing a wide array of chemicals. The integration of various bioinformatics tools and databases with the UISS framework enables a high degree of predictive accuracy, allowing for the identification of potential sensitizers and a deeper understanding of their immune-modulating effects. As this approach is highly adaptable and scalable, it can easily be extended to accommodate new data and more complex biological interactions, ensuring its relevance as toxicological knowledge evolves. By leveraging cutting-edge bioinformatics and computational tools, this approach promotes innovation in the field of toxicology and immunology, encouraging the development of new methodologies and tools that can further enhance the predictive accuracy and applicability of in silico models. Ultimately, the ability to accurately predict chemical sensitization and immune responses contributes to improved public health safety by ensuring that potentially harmful chemicals are identified and assessed more effectively, reducing the risk of allergic reactions and other adverse health effects.
Future plans include expanding the range of chemicals tested to enhance the models’ predictive accuracy and applicability. This expansion aims to improve chemical safety and support the development of more sustainable and ethical testing methods. Additionally, refining these models will help elucidate the mechanisms of chemical sensitization, leading to better-targeted interventions to prevent adverse reactions.
In conclusion, our study represents a crucial advancement in the modernization of toxicological assessments via computational science. The capacity to predict chemical sensitization with high accuracy, reduce dependency on animal testing, and deepen the understanding of immune mechanisms delivers substantial ethical, regulatory, and scientific benefits. This novel approach paves the way for safer, faster, and more cost-effective chemical evaluations, positioning it as a key player in the future of toxicology. Continued research, validation, and collaboration will be essential in further refining and expanding the capabilities of this method, contributing to more innovative and responsible toxicological practices worldwide.
Key Points
Innovative protocol development: A sophisticated eight-step bioinformatic protocol integrates data from multiple sources (PubChem, Protein Data Bank, AlphaFold) and employs agent-based modelling to predict chemical sensitization effects accurately.
Nonanimal testing approach: This study presents a computational framework that reduces reliance on animal testing by predicting the allergenic potential of chemicals using advanced simulation tools like the UISS.
Case study validation: The protocol’s efficacy is demonstrated through case studies with DNCB and TMA, well-known skin and respiratory sensitizers, respectively, showcasing its precision in distinguishing between different types of chemical allergens.
Implications for safety assessments: The successful application of this protocol can transform chemical safety evaluations, offering a more ethical, economical, and environmentally friendly alternative to traditional testing methods.
Acknowledgements
F.P., G.R., and E.C. were supported through an expert contract from the European Commission, Joint Research Centre (JRC). The information and views set out in this article are those of the authors and do not necessarily reflect the official opinion of the European Commission. Neither the European Commission institutions and bodies nor any person acting on its behalf may be held responsible for the use which may be made of the information contained therein.
Conflict of interest: None declared.
Contributor Information
Giulia Russo, Department of Drug and Health Sciences, University of Catania, V.le A. Doria, 6, 95125 Catania (IT), Italy.
Elena Crispino, Department of Biomedical and Biotechnological Sciences, University of Catania, Via S. Sofia, 63, 95125 Catania (IT), Italy.
Silvia Casati, European Commission, Joint Research Centre (JRC), Via Enrico Fermi, 2749 - TP 123 21027 - Ispra (VA), Italy.
Emanuela Corsini, Department of Pharmacological and Biomolecular Sciences, Università degli studi di Milano, Via Balzaretti 9, 20133 Milano, Italy.
Andrew Worth, European Commission, Joint Research Centre (JRC), Via Enrico Fermi, 2749 - TP 123 21027 - Ispra (VA), Italy.
Francesco Pappalardo, Department of Drug and Health Sciences, University of Catania, V.le A. Doria, 6, 95125 Catania (IT), Italy.
Funding
None declared.
References
- 1. Martin SF. New concepts in cutaneous allergy. Contact Dermatitis 2015;72:2–10. 10.1111/cod.12311. [DOI] [PubMed] [Google Scholar]
- 2. Alinaghi F, Bennike NH, Egeberg A. et al. . Prevalence of contact allergy in the general population: a systematic review and meta-analysis. Contact Dermatitis 2019;80:77–85. 10.1111/cod.13119. [DOI] [PubMed] [Google Scholar]
- 3. Xu Z, Zeng Q, Yang D. et al. . Mathematical Modelling Indicates Th-Cell Targeted Antibody-Dependent Cellular Cytotoxic Is a Crucial Obstacle Hurdling HIV Vaccine Development 2024.
- 4. OECD . The adverse outcome pathway for skin sensitisation initiated by covalent binding to proteins. Organisation for Economic Cooperation and Development, 2014.
- 5. Landsteiner K, Jacobs J. Studies on the sensitization of animals with simple chemical compounds. J Exp Med 1935;61:643–56. 10.1084/jem.61.5.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Catalona WJ, Taylor PT, Rabson AS. et al. . A method for dinitrochlorobenzene contact sensitization: a clinicopathological study. N Engl J Med 1972;286:399–402. 10.1056/NEJM197202242860804. [DOI] [PubMed] [Google Scholar]
- 7. Sadekar N, Boisleve F, Dekant W. et al. . Identifying a reference list of respiratory sensitizers for the evaluation of novel approaches to study respiratory sensitization. Crit Rev Toxicol 2021;51:792–804. 10.1080/10408444.2021.2024142. [DOI] [PubMed] [Google Scholar]
- 8. Kimber I, Dearman RJ, Basketter DA. et al. . Chemical respiratory allergy: reverse engineering an adverse outcome pathway. Toxicology 2014;318:32–9. 10.1016/j.tox.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 9. Lalko JF, Kimber I, Gerberick GF. et al. . The direct peptide reactivity assay: selectivity of chemical respiratory allergens. Toxicol Sci 2012;129:421–31. 10.1093/toxsci/kfs205. [DOI] [PubMed] [Google Scholar]
- 10. Buehler EV. Delayed contact hypersensitivity in the guinea pig. Arch Dermatol 1965;91:171. 10.1001/archderm.1965.01600080079017. [DOI] [PubMed] [Google Scholar]
- 11. Pistollato F, Madia F, Corvi R. et al. . Current EU regulatory requirements for the assessment of chemicals and cosmetic products: challenges and opportunities for introducing new approach methodologies. Arch Toxicol 2021;95:1867–97. 10.1007/s00204-021-03034-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Corsini E, Engin AB, Neagu M. et al. . Chemical-induced contact allergy: from mechanistic understanding to risk prevention. Arch Toxicol 2018;92:3031–50. 10.1007/s00204-018-2283-z. [DOI] [PubMed] [Google Scholar]
- 13. Casati S, Asturiol D, Browne P. et al. . Standardisation and international adoption of defined approaches for skin sensitisation. Front Toxicol 2022;4:943152. 10.3389/ftox.2022.943152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Smith CK, Hotchkiss SAM. Allergic Contact Dermatitis: Chemical and Metabolic Mechanisms 2001.
- 15. Kimber I, Poole A, Basketter DA. Skin and respiratory chemical allergy: confluence and divergence in a hybrid adverse outcome pathway. Toxicol Res 2018;7:586–605. 10.1039/c7tx00272f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dearman RJ, Warbrick EV, Skinner R. et al. . Cytokine fingerprinting of chemical allergens: species comparisons and statistical analyses. Food Chem Toxicol 2002;40:1881–92. 10.1016/S0278-6915(02)00179-5. [DOI] [PubMed] [Google Scholar]
- 17. Obaidullah AJ, Alanazi MM, Alsaif NA. et al. . Deeper insights on Cnesmone javanica Blume leaves extract: chemical profiles, biological attributes, network pharmacology and molecular docking. Plants (Basel) 2021;10:728. 10.3390/plants10040728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Paul A, Das T, MdHU C. et al. . Anthelmintic activity of pineapple: in silico molecular docking and molecular dynamics simulation. 2024.
- 19. Rahman J, Tareq AM, Hossain MM. et al. . Biological evaluation, DFT calculations and molecular docking studies on the antidepressant and cytotoxicity activities of Cycas pectinata Buch.-Ham. compounds. Pharmaceuticals (Basel) 2020;13:232. 10.3390/ph13090232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pappalardo F, Russo G, Corsini E. et al. . Translatability and transferability of in silico models: context of use switching to predict the effects of environmental chemicals on the immune system. Comput Struct Biotechnol J 2022;20:1764–77. 10.1016/j.csbj.2022.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dearman RJ, Basketter DA, Kimber I. Characterization of chemical allergens as a function of divergent cytokine secretion profiles induced in mice. Toxicol Appl Pharmacol 1996;138:308–16. 10.1006/taap.1996.0129. [DOI] [PubMed] [Google Scholar]
- 22. Aronica MA, Mora AL, Mitchell DB. et al. . Preferential role for NF-kappa B/Rel signaling in the type 1 but not type 2 T cell-dependent immune response in vivo. J Immunol 1999;163:5116–24. [PubMed] [Google Scholar]
- 23. Farraj AK, Harkema JR, Kaminski NE. Topical application versus intranasal instillation: a qualitative comparison of the effect of the route of sensitization on trimellitic anhydride–induced allergic rhinitis in A/J mice. Toxicol Sci 2006;92:321–8. 10.1093/toxsci/kfj191. [DOI] [PubMed] [Google Scholar]
- 24. Plitnick LM, Loveless SE, Ladics GS. et al. . Cytokine profiling for chemical sensitizers: application of the ribonuclease protection assay and effect of dose. Toxicol Appl Pharmacol 2002;179:145–54. 10.1006/taap.2002.9370. [DOI] [PubMed] [Google Scholar]
- 25. Russo G, Crispino E, Corsini E. et al. . Computational modelling and simulation for immunotoxicity prediction induced by skin sensitisers. Comput Struct Biotechnol J 2022;20:6172–81. 10.1016/j.csbj.2022.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hopkins JE, Naisbitt DJ, Kitteringham NR. et al. . Selective haptenation of cellular or extracellular protein by chemical allergens: association with cytokine polarization. Chem Res Toxicol 2005;18:375–81. 10.1021/tx049688. [DOI] [PubMed] [Google Scholar]
- 27. Wang H, Zhang J, Lu Z. et al. . Identification of potential therapeutic targets and mechanisms of COVID-19 through network analysis and screening of chemicals and herbal ingredients. Brief Bioinform 2022;23:bbab373. 10.1093/bib/bbab373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol 2010;11:373–84. 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 29. Bieber T. Atopic dermatitis. N Engl J Med 2008;358:1483–94. 10.1056/NEJMra074081. [DOI] [PubMed] [Google Scholar]
- 30. O’Neill LAJ, Bowie AG. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat Rev Immunol 2007;7:353–64. 10.1038/nri2079. [DOI] [PubMed] [Google Scholar]
- 31. Guilliams M, Lambrecht BN, Hammad H. Division of labor between lung dendritic cells and macrophages in the defense against pulmonary infections. Mucosal Immunol 2013;6:464–73. 10.1038/mi.2013.14. [DOI] [PubMed] [Google Scholar]
- 32. Lambrecht BN, Hammad H. Biology of lung dendritic cells at the origin of asthma. Immunity 2009;31:412–24. 10.1016/j.immuni.2009.08.008. [DOI] [PubMed] [Google Scholar]
- 33. Jumper J, Evans R, Pritzel A. et al. . Highly accurate protein structure prediction with AlphaFold. Nature 2021;596:583–9. 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pettersen EF, Goddard TD, Huang CC. et al. . UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 2004;25:1605–12. 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 35. Zhang W, Bell EW, Yin M. et al. . EDock: blind protein–ligand docking by replica-exchange Monte Carlo simulation. J Chem 2020;12:37. 10.1186/s13321-020-00440-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yan Y, Tao H, He J. et al. . The HDOCK server for integrated protein–protein docking. Nat Protoc 2020;15:1829–52. 10.1038/s41596-020-0312-x. [DOI] [PubMed] [Google Scholar]
- 37. Ponomarenko J, Bui H-H, Li W. et al. . ElliPro: a new structure-based tool for the prediction of antibody epitopes. BMC Bioinformatics 2008;9:514. 10.1186/1471-2105-9-514. [DOI] [PMC free article] [PubMed] [Google Scholar]