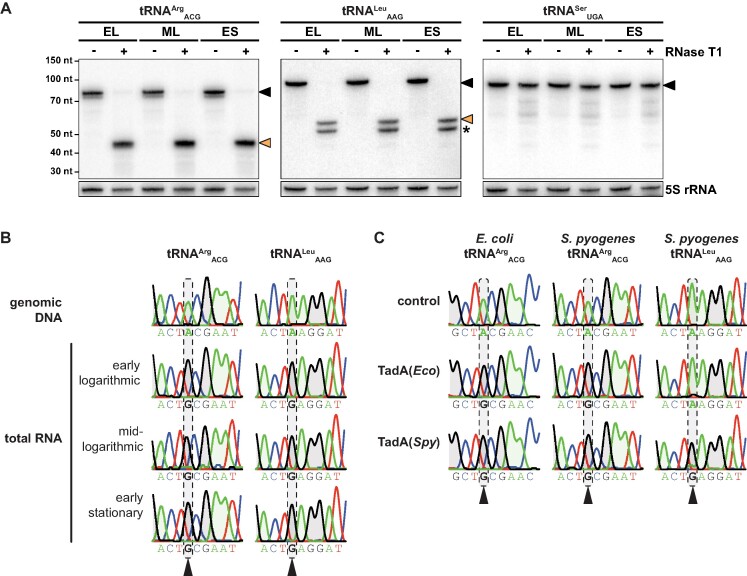
Figure 1.
A34-to-I34 editing of S. pyogenes tRNAs in vivo and in vitro. (A) Expression and editing status of the two A34-tRNAs of S. pyogenes and of a non-A34 control tRNA were analysed by RNase T1 treatment of glyoxal/borate-protected total RNA and subsequent Northern blotting against tRNA 3′ ends. Full-length tRNAs are marked with a black arrow and inosine-dependent cleavage products with an orange arrow. Expected sizes are 77 nt versus 42 nt for tRNAArgACG, 89 nt versus 54 nt for tRNALeuAAG, and 93 nt for the negative control tRNASerUGA (no I34 modification). An additional cleavage fragment for tRNALeuAAG, likely due to the presence of another tRNA modification that inhibits the formation of an RNase T1-resistant guanosine adduct, is marked by an asterisk. 5S rRNA was used as loading control. EL: early logarithmic; ML: mid-logarithmic; ES: early stationary. (B) The editing status of the two A34-tRNAs of S. pyogenes was further examined by Sanger sequencing at three different growth phases (three lower panels) and compared with the genomic DNA control (upper panel). (C)In vitro transcribed A34-tRNAs from E. coli (tRNAArgACG only) and S. pyogenes (tRNAArgACG and tRNALeuAAG) were deaminated in vitro with recombinant TadA from both species or with buffer only as control. The Sanger chromatograms in (B) and (C) correspond to tRNA positions 31–39 with the editing position 34 highlighted by grey dashed boxes. Nucleotide sequences are shown below each chromatogram. Sequencing traces of adenosine and guanosine are shaded in green and grey, respectively, for better visualisation.