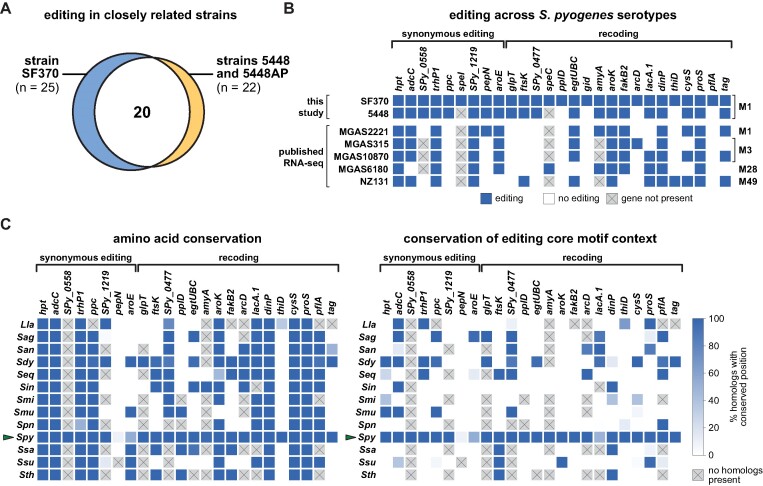
Figure 3.
Conservation of mRNA editing in S. pyogenes and other Streptococcaceae. (A) Venn diagram of editing target genes in S. pyogenes strain 5448 and hypervirulent strain 5448AP (identical in both strains) compared with strain SF370. (B) Editing signatures in editing target genes in RNA-seq datasets from this study (upper rows) and publicly available RNAseq datasets (lower rows) of various S. pyogenes strains (names on the left) and M serotypes (indicated on the right). (C) Conservation of the amino acid (left) and TAMG core consensus context (right) in homologs of S. pyogenes SF370 editing target genes in selected Streptococcaceae family members. The fraction of homologs within each species (left) with identical amino acid and editable codons, respectively, is depicted for each editing target gene (top) as a heatmap. Synonymously edited and recoded target genes are grouped accordingly. S. pyogenes is highlighted by a green arrow. For homologs of SPy_0477, editing was also considered possible in the case of the TATG sequence, given the native editing context of the gene in S. pyogenes SF370. Species: Lactococcus lactis (Lla), Streptococcus agalactiae (Sag), S. anginosus (San), S. dysgalactiae (Sdy), S. equi (Seq), S. iniae (Sin), S. mitis (Smi), S. mutans (Smu), S. pneumoniae (Spn), S. pyogenes (Spy), S. salivarius (Ssa), S. suis (Ssu) and S. thermophilus (Sth).