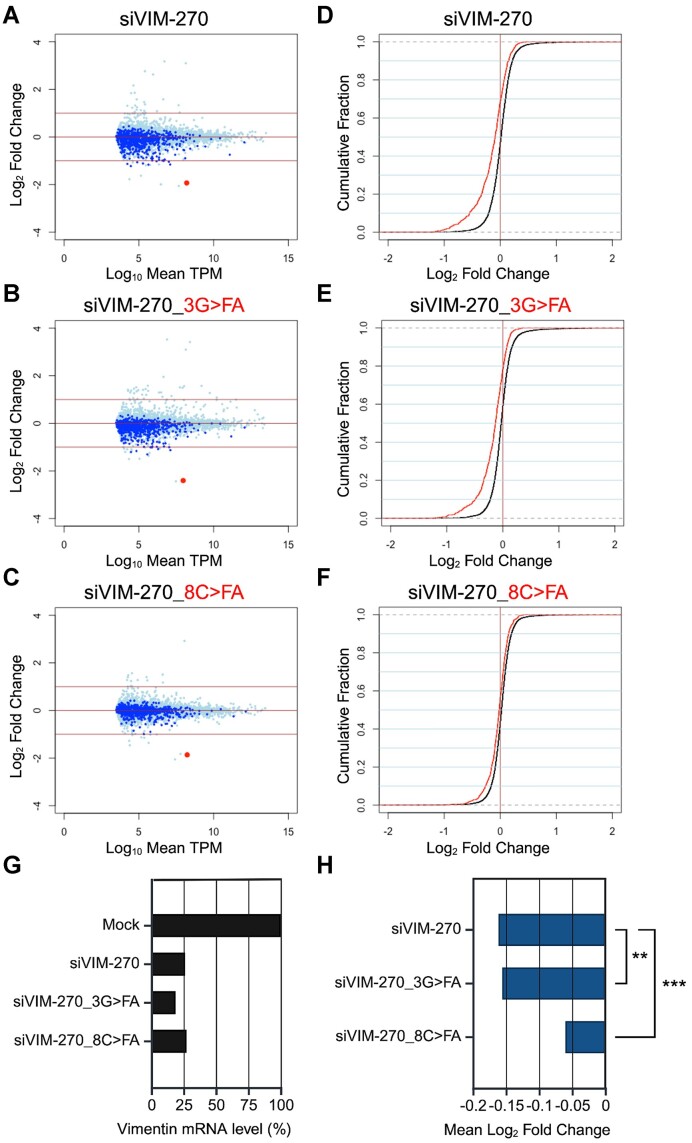
Figure 5.
RNA-seq analysis of on-target and off-target effects of siVIM-270 with 2′-formamido modifications (A-C). MA plots showing log2 fold changes (vertical axis) and average log10 transcripts per million (TPM) (horizontal axis) of the expression levels of transcripts from cells transfected with siVIM-270 and its variants versus those from mock-transfected cells. Dark blue dots represent 676 transcripts with the siVIM-270 SM sequence (off-target transcripts) in their 3′ UTRs, while light blue dots represent 8076 other transcripts without the SM sequence (not off-target transcripts). (D-F) Cumulative distributions of transcripts showing log2 fold changes (horizontal axis) and cumulative fractions (vertical axis). Red and black lines show the results for off-target and non-off-target transcripts, respectively. (G) VIM mRNA levels calculated by TPM compared to mock. (H) Seed-dependent off-target effects shown by mean log2 fold change for unmodified siVIM-270 and 2′-formamido-modified siVIM-270s at positions 3 or 8. Statistical significance for each group was determined by comparing the cumulative distribution differences of siRNA SM transcripts using the Wilcoxon rank-sum test (**P < 0.01, ***P < 0.001).