Figure 3: EVN arrests ribosomes at sites encoding specific tripeptide motifs.
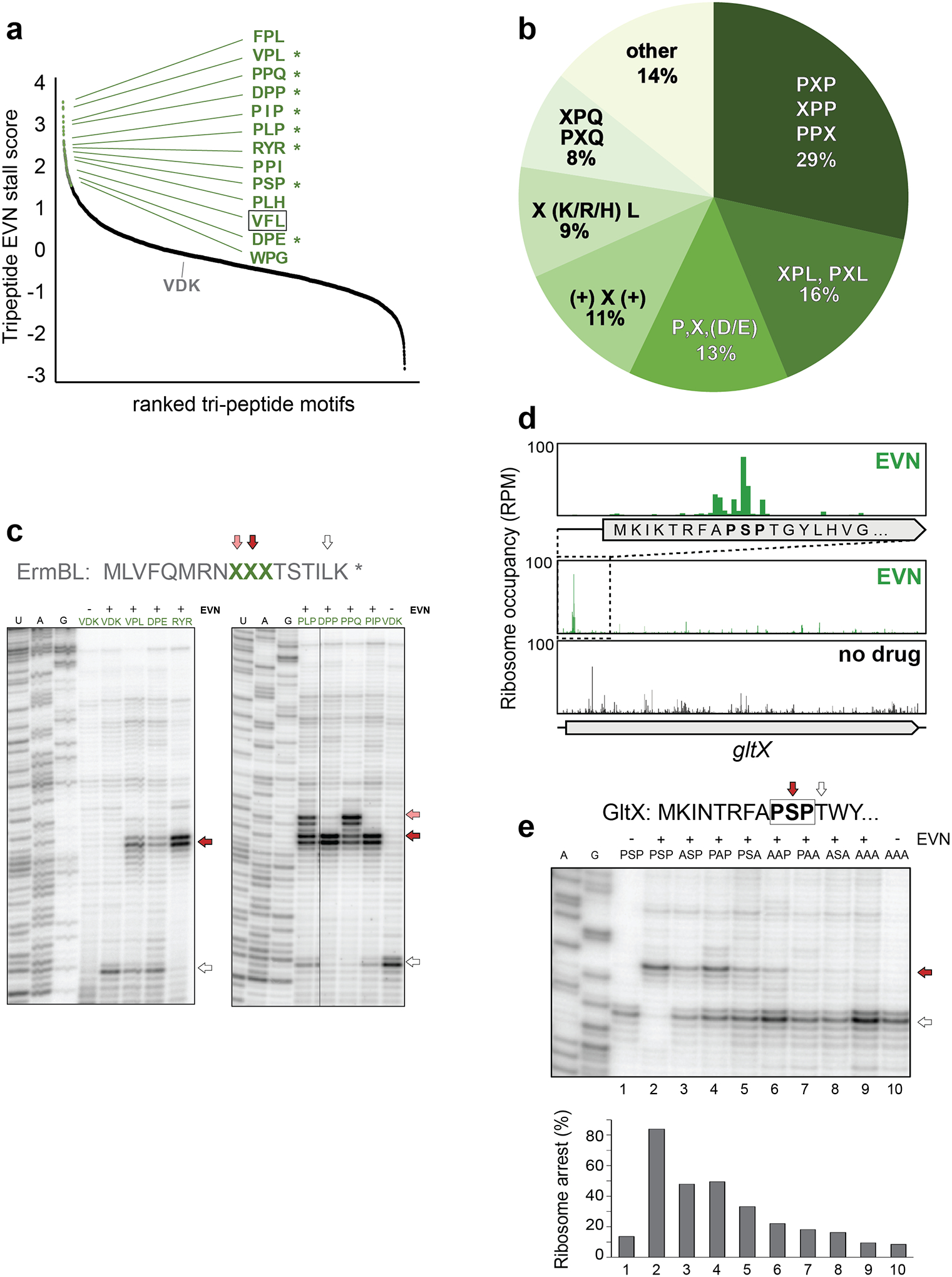
a, Plot of EVN stall scores for tripeptides motifs. Tripeptides with stall score ≥ 2 SD from the mean of all motifs are indicated with green dots. Representative tripeptide sequences are shown. Sequences tested in toeprinting experiments (panels c and e) are marked by asterisks. The tripeptide sequence VFL, which is found in the EmtAL leader peptide (see Fig. 4), is boxed. Ranking placement of the non-stalling VDK peptide that was mutated in the ermBL template (panel c) is indicated. b, Relative occurrence of the most prevalent tripeptide motifs at the sites of strongest EVN action (n = 98). c, In vitro toeprinting for ribosome stalling on ermBL templates where the segment encoding the original Val9Asp10Lys11 sequence of the ORF was swapped with sequences encoding specific EVN-arrest tripeptide motifs (among those shown in panels a and b). Translation arrest by EVN at the introduced sites are indicated with red and pink arrows. The white arrow shows the ribosomes that, after escaping arrest at the introduced tripeptide motifs, stall at Leu16-UUG codon of ermBL (see Fig. 1a). Lanes corresponding to samples containing no antibiotic are labeled with “-”. Sequencing reactions lanes are labeled as U, A, G. d, Ribosomal density profile of the gltX gene in control or EVN treated cells. The inset shows EVN-mediated of ribosome accumulation at the early gltX PXP motif Pro8Ser9Pro10. e, Toeprinting analysis of EVN-mediated arrest at the modified gltX template (see Extended Data Fig. 6a) with the original or mutated Pro8Ser9Pro10 motif. Red arrow indicates ribosomes with EVN stalled at codon 10. Ribosomes that escape EVN arrest become stalled at Thr12 codon (white arrow) preceding the introduced Trp13 codon, because reactions are depleted from Trp-tRNA by the addition of the Trp-RS inhibitor indolmycin. Lanes of samples with no EVN are marked with “-”. Sequencing reactions lanes are labeled as A, G. Toeprinting analysis of ribosome stalling by EVN at the unmodified gltX template is shown in Extended Data Fig 6a. Representative gels from at least two independent experiments are shown in panels c and e.
