Figure 4: Programmed EVN-mediated translation arrest may control expression of orthosomycin-resistance genes.
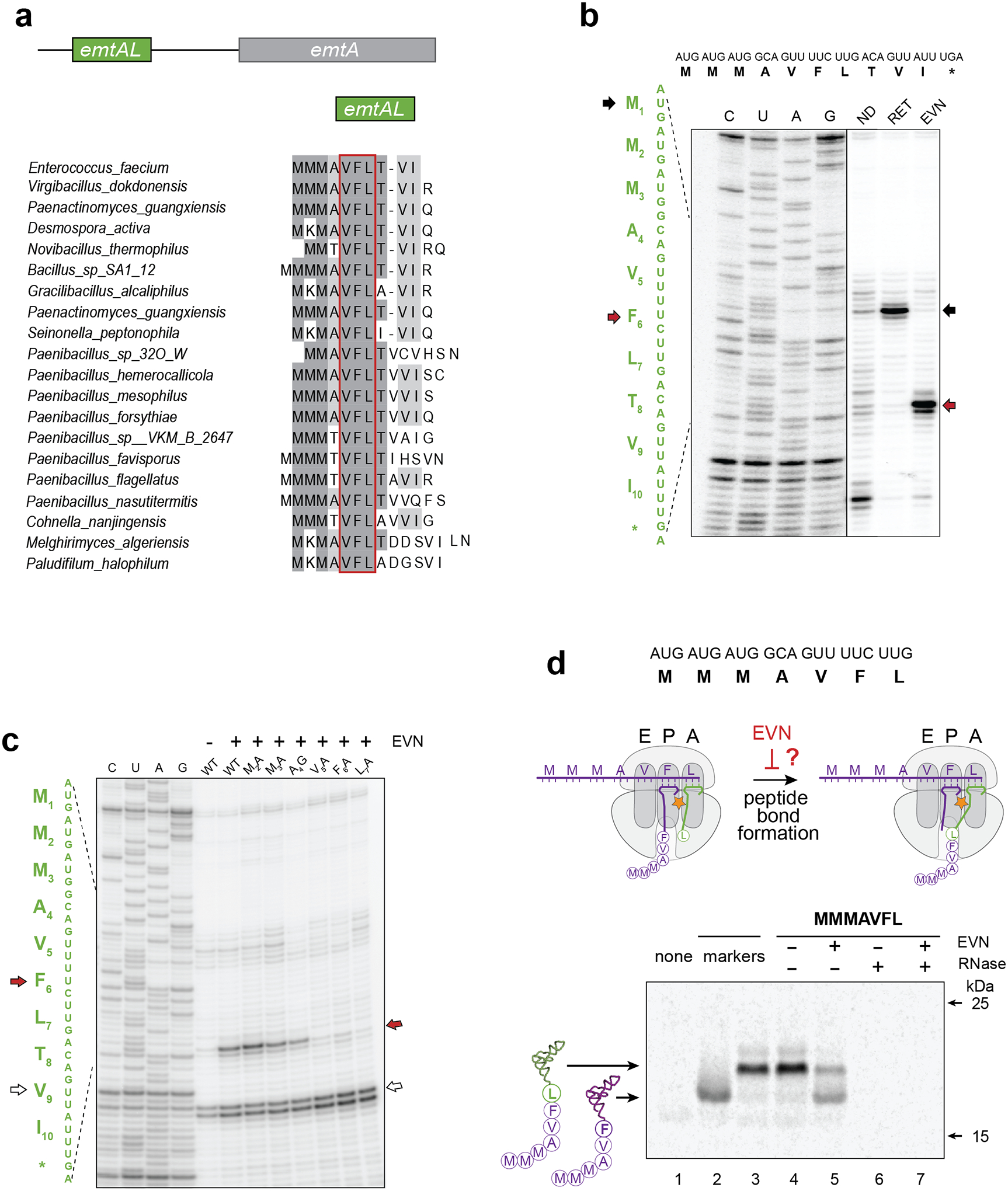
a, A conserved leader small ORF (that we named emtAL) precedes the orthosomycin-resistance gene emtA. The amino acid sequences encoded by emtAL ORFs of different bacterial species is shown. The VFL sequence (boxed in red) present in all the identified emtAL leader ORFs, is one of the tripeptide motifs associated with the strongest EVN arrest sites (see Fig. 3 a–b). b, Toeprinting analysis of ribosome arrest induced by EVN during in vitro translation of the emtAL ORF of Enterococcus faecium. Inclusion of the initiation inhibitor retapamulin20 (sample labeled RET) helped identified the first of the three consecutive AUG codons as the start codon of the ORF (marked with black arrows). Red arrows indicate ribosomes stalled by EVN at the Phe6 codon of the emtAL VFL sequence. c, Toeprinting analysis of EVN-mediated ribosome stalling in mutant emtAL ORF where codons 2–7 of the wt emtAL were individually substituted by Ala or Gly codons. Red arrow indicates ribosomes stalled by EVN at codon Phe6 of the ORFs. White arrow shows ribosomes trapped at Val9 codon because reactions lacked Ile-tRNA due to the presence of Ile-RS inhibitor mupirocin. Sequencing reactions are shown. d, Testing whether EVN inhibits peptide bond formation. Top: sequence of the truncated emtAL template, encoding the MMMAVFL peptide and lacking a stop codon, used in the experiment. Middle: cartoon representation of the experimental scheme. Inhibition of peptide bond formation by EVN would lead to the appearance of MMMAVF-tRNAPhe. Bottom: Gel electrophoresis of the in vitro translation products. Reactions contained [14C]-Phe and were carried without or with EVN (500 μg/mL). Reaction on lane 1 (‘none’) contained no template. Mobility markers (lanes 2 and 3) were prepared by translating non-stop templates encoding the MMMAVF or MMMAVFL peptides. Samples in lanes 6 and 7 were treated with RNase I; the resulting small peptides (MMMAVF and MMMAVFL) are not retained in the gel. Electrophoretic mobility of molecular weight protein markers (of 25 and 15 kDa) is indicated. The Coomassie-stained gel is shown in the Source Data file. Representative gels from at least two independent experiments are shown in panels b, c and d.
