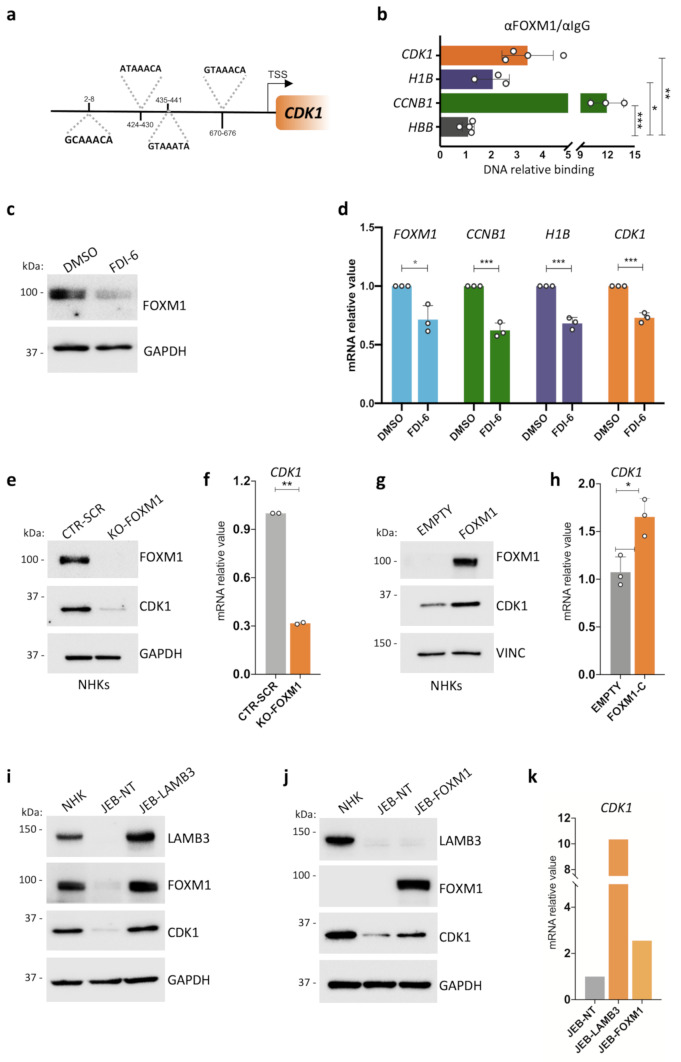
Fig. 2.
A feedforward loop between FOXM1 and CDK1 in human primary epidermal cells. (a) Schematic representation of FOXM1 predicted binding sites on the CDK1 promoter. (b) ChIP-ddPCR showing FOXM1 binding to the indicated sites in NHK. Relative DNA binding was calculated as a fraction of input and normalized with the negative region (HBB, see Suppl. Table 5) arbitrarily set to 1, N = 3 independent experiments. *P < 0.05, **P < 0.01 ***P < 0.001 (Student t-test). (c) Western analysis of total cell extracts derived from NHK cultures treated with 30 μM FDI-6 or DMSO as control. Molecular weight indicators are shown. (d) qRT-PCR quantification of the mRNA levels of FOXM1, CCNB1, H1B, and CDK1 on cells derived from DMSO- or FDI-6 -treated NHK cultures. Expression levels were normalized per GAPDH and given relative to the control (DMSO) arbitrarily set to 1. Data are presented as mean +/− SD of N = 3 different independent biological replicates, *P < 0.05, ***P < 0.001 (Student t-test). (e) Western analysis of total cell extracts from cultures generated by primary NHKs edited with a scrambled gRNA (CTR-SCR) as control or with a gRNA specific for FOXM1 (KO-FOXM1). Molecular weight indicators are shown. (f) qRT-PCR quantification of the mRNA levels of CDK1 on cells derived from gRNA scrambled- (CTR-SCR) or FOXM1-edited (KO-FOXM1) NHK. Expression levels were normalized per GAPDH. Data are presented as mean +/− SD of N = 2 biological replicates, **P < 0.01 (Student t-test). (g) Western analysis on total cell extracts derived from FOXM1- (FOXM1) or empty backbone-transduced (EMPTY) NHK cultures. FOXM1 endogenous level in NHK-empty is not detectable due to technical reasons. Molecular weight indicators are shown. (h) qRT-PCR quantification of the mRNA levels of CDK1 on cells derived from FOXM1- (FOXM1) or empty backbone-transduced (EMPTY) NHK cultures. Expression levels were normalized per GAPDH. Data are presented as mean +/− SD of N = 3 different independent biological replicates, *P < 0.05 (Student t-test). (i) Western analysis on total cell extracts derived from NHK, untransduced (JEB-NT) and LAMB3-transduced (JEB-LAMB3) JEB-derived primary keratinocyte cultures. Molecular weight indicators are shown. (j) Western analysis on total cell extracts derived from untransduced (JEB-NT) and FOXM1-transduced (JEB-FOXM1) JEB-derived primary keratinocyte cultures. Molecular weight indicators are shown. (k) qRT-PCR quantification of the mRNA levels of CDK1 on cells derived from untransduced (JEB-NT), FOXM1- (JEB-FOXM1), or LAMB3-transduced (JEB-LAMB3) JEB-derived primary keratinocyte culture. Expression levels are normalized per GAPDH. Data from one available patient are shown