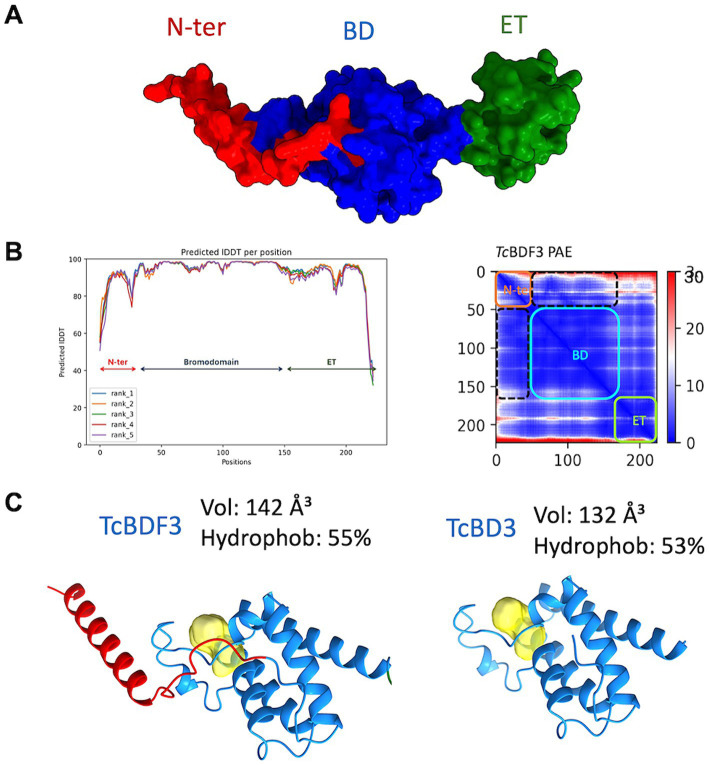
Figure 3.
(A) Structure of TcBDF3 predicted by AlphaFold2 (surface representation). N-ter, N-terminal α-helix (red); BD, Bromodomain (blue); ET, extraterminal domain (green). (B) Predicted IDDT per position for the five models obtained and Predicted Aligned Error (PAE) plot obtained from AF2 prediction of TcBDF3 structure. Dash lines indicate the interface between the BD and the N-tern α-helix in the PAE plot. In the plot blue color indicates lower PAE, which means the best prediction of relative ubication of the amino acids. (C) Volume and hydrophobicity of BD cavity from full-length TcBDF3 and isolated bromodomain (TcBD3) calculated with CAVIAR.