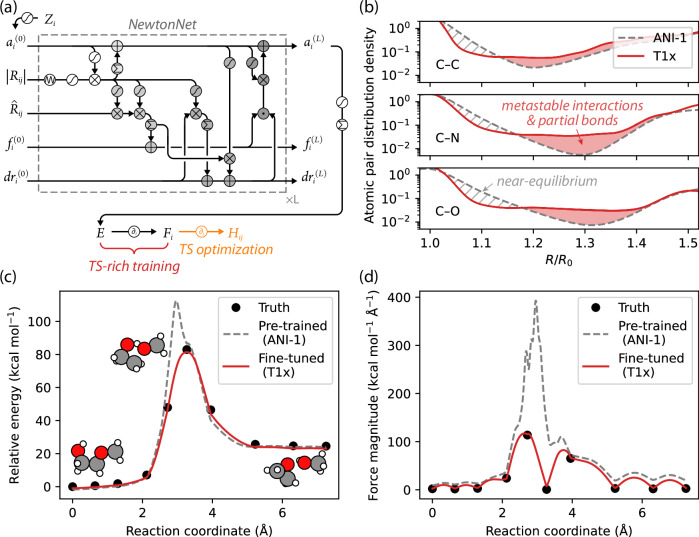
Fig. 1. The NewtonNet model and its performance on the ANI-1 and T1x data sets.
a The equivariant message-passing neural network designed for 3D molecular graphs with nodes {Zi} and edges {Rij} to predict molecular energies E and atomic forces Fi32. In this work, we further differentiate the network to derive Hij for TS optimization tasks. b The distribution of atomic pairwise distances, R, relative to equilibrium bond distances, R0, among datasets we used for training, where the T1x data set provides more data in the TS region, and further augmented with the ANI-1x data to add data corresponding to bond compression. The predicted (c) potential energies and (d) forces along the reaction coordinate for an unseen reaction for the pre-trained and fine-tuned model. A comprehensive statistical analysis of energy and force prediction errors along the reaction coordinates for 1248 unseen test reactions is summarized in Supplementary Fig. S1 for the pre-trained and fine-tuned models. Details of the training protocols are described in Methods and Supplementary Fig. S2 and S3. TS: transition state. Source data for this figure are provided with this paper.