Abstract
Purpose:
To identify predictive factors of nivolumab sensitivity, peripheral blood NKs and regulatory T-cell (Treg) were evaluated in patients with metastatic renal cell carcinoma (mRCC) enrolled in the REVOLUTION trial.
Experimental Design:
Fifty-seven mRCCs being treated with nivolumab, as at least second-line of therapy, and 62 healthy donors were longitudinally evaluated (0–1–3–6–12 months) for peripheral NKs and Tregs, phenotype, and function. Multivariable logistic regression was conducted to identify the independent predictors. The 0.632+ internal cross-validation was used to avoid overfitting. The best cutoff value based on a 3-month clinical response was applied to progression-free survival (PFS) and overall survival (OS). Kaplan–Meier curves for PFS and OS were produced.
Results:
At pretreatment, mRCCs displayed high frequency of NKp46+NKs, NKp30+NKs, KIR2DL1+NKs, KIR2DL2/DL3+NKs, and PD1+NKs with reduced NK degranulation as well as high frequency of Tregs, PD1+Tregs, Helios+Tregs, and ENTPD1+Tregs. Responder patients, identified as a clinical response after 3 months of treatment, presented at pretreatment significantly low CD3+, high KIR2DL2/DL3+NKs, high PD1+Tregs, and high Helios+Tregs. Upon multivariate analysis, only KIR2DL2/DL3NKs and Helios+Tregs held as independent predictors of nivolumab responsiveness. The KIR2DL2/DL3+NKs >35.3% identified patients with longer OS, whereas the Helios+Tregs >34.3% displayed significantly longer PFS. After 1-month of nivolumab, responder patients showed low CD3+, high NKs, KIR2DL2/DL3+NKs, and ICOS+Tregs. Among these subpopulations, CD3+ and KIR2DL2/DL3+NKs held as independent predictors of nivolumab efficacy. Low CD3+ (≤71%) was significantly associated with longer PFS, whereas high KIR2DL2/DL3+NKs (>23.3%) were associated with both PFS and OS.
Conclusions:
Pretreatment evaluation of Helios+Tregs/KIR2DL2/DL3+NKs and 1-month posttreatment CD3+/ KIR2DL2/DL3+NKs will predict nivolumab response in mRCCs.
Translational Relevance.
Although immune checkpoint inhibitors represent the core of first-line therapy in advanced/metastatic renal cancer, only a subset of patients benefit from treatment. Thus, biomarkers predictive of response are eagerly awaited. In this study, baseline peripheral KIR2DL2/DL3+NKs and Helios+Tregs or early posttreatment values of CD3+ and KIR2DL2/DL3+NKs represent early, feasible, predictive biomarkers of nivolumab responsiveness in patients with mRCC. Easy integration of blood testing might significantly improve metastatic renal cancer patient management.
Introduction
Kidney cancer is the ninth malignancy among males and 14th among females (1) with more than 400,000 new cases diagnosed worldwide each year (2). Clear cell renal cell carcinoma is the most common histotype accounting for more than 80% of cases (3). Von Hippel–Lindau (VHL) inactivation is detected in the majority (57%–80%) of clear cell renal cell carcinoma tumors (4) and additional common genetic alterations include PBRM1, BAP1, SETD2, TP53, and KDM5C mutations (5). Approximately 25% of patients present with advanced-stage disease, whereas 30% will develop distant metastases during follow-up (6, 7). Standard therapies for advanced/metastatic renal cell carcinoma (mRCC) rely on combinations of tyrosine kinase inhibitors (axitinib and cabozantinib) and immune checkpoint inhibitors (ICI; nivolumab and ipilimumab) or ICI combinations (8). Nevertheless, only a subset of patients will benefit from ICI treatments (9), thus reliable biomarkers of response represent unmet needs (10). Tissue biomarkers, such as tumor mutational burden and programmed death ligand-1 (PDL1), seem less informative in RCC than in melanoma or non–small cell lung cancer (NSCLC; refs. 11, 12). Although RCCs display a relatively modest rate of mutations [1.42 mutations per megabase (mut/mb)] (13, 14), it is considered immunogenic because of spontaneous regressions (13), efficacy of IL2 (14), and response to ICIs (9, 15, 16).
The tumor immune microenvironment (TIME) is characterized by T cells, B cells, NKs, dendritic cells (DC), myeloid-derived suppressor cells plus cytokines, such as IL10 and TGFβ, which suppress immune cell activation and promote the differentiation of regulatory T cells (Treg; ref. 17). Pretreatment expanded CD8+ T-cell clones have been detected in patients whose mRCC responded to nivolumab; notably, the binding of nivolumab induced granzyme B, suggesting that CD8+ T cells have a cytotoxic potential and are likely tumor-reactive (18, 19). However, the intricate and diverse RCC-TIME gives rise to substantial heterogeneity also in response to immunotherapy (20). Tregs and NKs play a crucial role in TIME in which Tregs impair NK’s effector functions (21). The Treg impairment of NK function was described in patients with NSCLC in which circulating NKs progressively increase concomitantly with a decrease in Tregs in Responder patients to nivolumab (22). Anti-CTLA4 reduces Tregs and Treg-mediated NK inhibition in patients with head and neck cancer (23). TGFβ membrane-bound Tregs and Tregs–IL2, IL10 impaired NK activity by downregulating the activating receptor NKG2D on NK cells (24, 25). Together these findings suggest that Tregs can limit NK-cell activity through secreted cytokines and cell–cell contact.
NKs recognize tumor cells through a large repertoire of germline-encoded receptors with activating (DNAM-1, NKG2D, 2B4, NKG2C, NKp30, NKp44, NKp46, and NKp80; ref. 26) or inhibitory properties like CD85j (ILT2), KIR, NKG2A, and TIGIT (26, 27). The sum of activating and inhibiting signals determines the final NK-cell activation status and cytotoxic capacity (28). The role of PD1 on NKs is controversial, as PD1+NKs have been reported as both functionally defective (27, 29) or activated (30, 31) cells. A comparable controversy is the role of PD1 on Tregs. Evidence suggests that PD1 on Tregs enhances the FOXP3 expression and thereby increases their stability (32). The efficacy of anti-PD1 therapies was predicted by the balance of PD1 expression between CD8 and Treg cells, if PD1 is predominant on CD8, the PD1 blockade unleashes antitumor immune response, if PD1 is predominant on Tregs the PD1 blockade induces their activation and expansion (19, 33).
To investigate peripheral predictors of nivolumab sensitivity, we focused on the longitudinal evaluation of Tregs and NKs, phenotype, and function, hypothesizing that these features would mirror the TIME status.
Materials and Methods
Study design and patient enrollment
The REVOLUTION trial (NCT03891485) was a longitudinal, prospective, observational, multicentre study conducted between April 2016 and June 2022 in three centers, IRCCS-Napoli Pascale (Italy), Cancer Research Center of Lyon, Léon Bérard Center (France) and Val d’Hebron University Hospital (Spain). The study was conducted in accordance with the Declaration of Helsinki and approved by the Ethical Committee of the coordinating Institution (CE #21/16 OSS). The main inclusion criteria were as follows: (i) age of at least 18 years, (ii) pathologically confirmed mRCC, (iii) nivolumab treatment as second or following lines of therapy (from now patients with mRCC-REV or REV), (iv) signed informed consent. Eligible mRCC-REV patients received nivolumab at 3 mg/kg every 2 weeks until disease progression or unacceptable toxicity. The sample size of nivolumab-treated subjects was calculated on cell population variation in subjects with an objective response compared with subjects without an objective response. Thus, we estimated a priori that a sample size of 20 patients with response (R) and 37 with no response (NR; total n = 57) could achieve 80% power to reject the null hypothesis of equal means when the population mean difference is 0.8 (i.e., effect size) with a significance level (α) of 0.050 using a two-sided two-sample equal-variance t test. Herein, a posteriori, we report 33 R and 22 NR, proportions that hold the effect size.
Response assessment
Patients underwent clinical evaluation after 3 months of nivolumab treatment. Tumor response was assessed with CT scans and classified according to Response Evaluation Criteria in Solid Tumors v. 1.1. as complete response (CR), partial response (PR), stable disease (SD) or progressive disease (PD), and death (D). Patients were classified as follows: Rs comprising those with CR, PR, and SD with PFS ≥6 months (SD), or NRs comprising patients with SD <6 months, those with PD, and those who have died (D; refs. 34–36). Short-term nivolumab efficacy was evaluated by disease control rate (DCR) = (CR + PR + SD)/total cases × 100% and long-term efficacy was evaluated through PFS, the time from the first nivolumab administration to progression, and overall survival (OS), the time from the first nivolumab administration to death/last clinical evaluation. In case of death before disease progression, PFS was equal to OS time.
Peripheral blood collection
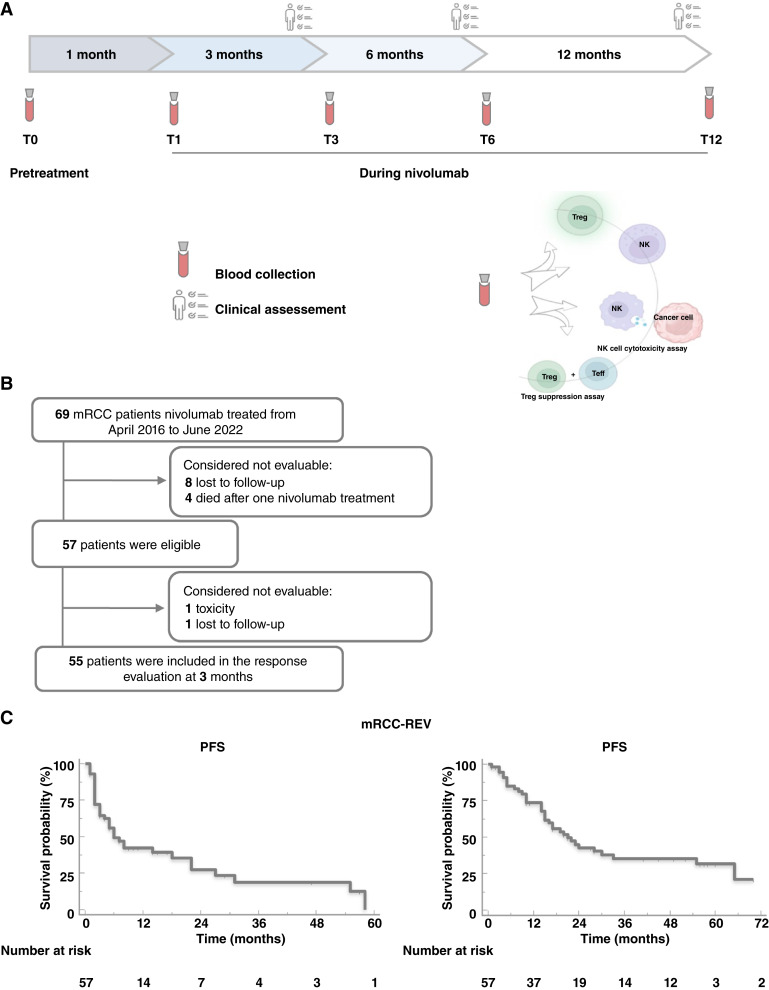
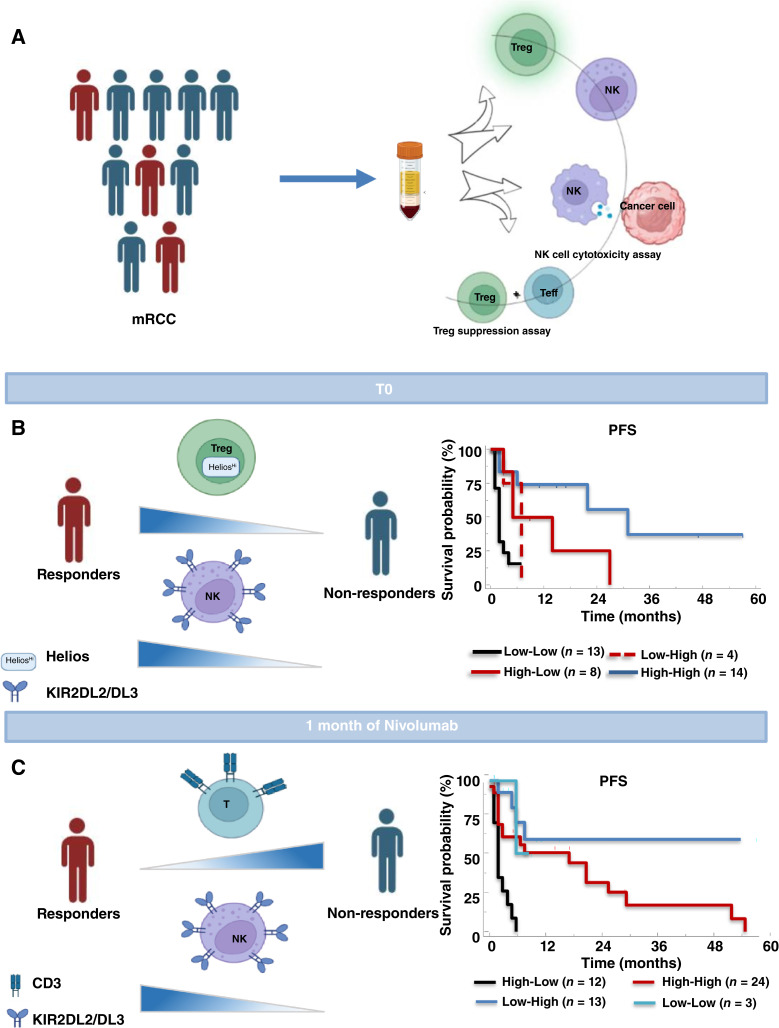
Heparinized peripheral blood (PB) samples (24 mL) were collected before starting nivolumab (REV, n = 69; baseline, T0), at 1 month (T1), 3 months (T3), 6 months (T6), and 12 months (T12) of treatment (Fig. 1). In addition, PB was collected from 62 healthy donors (HDs) at the Transfusion and Stem Cell Transplantation Unit-IRCCS-Naples. Peripheral blood mononuclear cells (PBMC) were isolated by Ficoll–Hypaque (GE Healthcare Bioscience Cytiva, Cat. #17-1440-02) density gradient following standardized protocols.
Figure 1.
Study design and Kaplan–Meier curves in patients with mRCC. A, Blood samples (24 mL) were collected before nivolumab (baseline, T0) and after 1 (T1), 3 (T3), 6 (T6), and 12 (T12) months of treatment. Clinical assessment every 3 months. B, Study enrollment flow chart. C, Kaplan–Meier curve for PFS and overall survival (OS) in 57 nivolumab-treated patients with mRCC.
Flow cytometry
Flow cytometry was performed on fresh PB samples on BD FACS Aria III flow cytometer. The optimal performance of the flow cytometer was measured daily with CS&T beads (BD Biosciences, Cat. # 655050), and the BD CompBeads particles (BD Biosciences, Cat. # 552843, RRID: AB_10051478) were used to compensate spectral overlap according to the manufacturer’s instructions. For surface markers, cells were incubated with antibodies for 30 minutes at 4°C, and washed with BD staining buffer (BSA; BD Biosciences, Cat. # 554657, RRID:AB_2869007). Intracellular antigens were assessed with a commercially available kit (BD Cytofix/Cytoperm; fixation and permeabilization [BD Biosciences, Cat. # 554655, RRID:AB_2869005; BD Biosciences, Cat. # 554723, RRID:AB_2869011]) according to the manufacturer’s instructions. Viability was analyzed using LIVE/DEAD cell stain (Invitrogen by ThermoFisher, Cat. #L34966). The list of antibodies used for immune cell phenotyping is detailed in Supplementary Table S1. The following populations were determined: NKs (CD3, CD56, CD16, NKp30, NKp44, NKp46, NKG2D, KIR2DL1, KIR2DL2/DL3, KLRB1, and PD1) and Tregs (CD4, CD25, CD127, FOXP3, CTLA4, PD1, ENTPD1, ICOS, Helios, and CXCR4). NKs and Tregs gating strategies were shown in Supplementary Fig. S1A and S1B, respectively. A minimum of 100,000 events for each sample were collected and data were analyzed using FacsDiva software 8.01 (BD FACSDiva Software RRID:SCR_001456).
NK-cell cytotoxicity assay
CD3−CD56+ NKs were isolated from fresh PBMCs by negative selection using a magnetic beads cell sorting system according to the manufacturer’s instructions (NK-cell Isolation Kit, human, Miltenyi Biotec, Cat. #130-092-657). NKs were stimulated with recombinant interleukin-2 (rIL2, 100 U/mL; Miltenyi Biotec, Cat. #130-097-742) for 18 hours in RPMI-1640 complete medium before performing cytotoxicity assays. CD107a (LAMP-1), markers of NK-cell functional activity, was evaluated on rIL2-activated NKs (effector cells) co-cultured with target K562 cells [RRID:CVCL_0004, obtained from the National Cancer Institute’s Developmental Therapeutics, program NCI DTP; the cells were routinely tested to ensure that they were mycoplasma free and authenticated based on short tandem repeats at Eurofins Genomics] at 10:1 effector/target ratio in the presence of PE anti-CD107a antibody for 4 hours at 37°C and 5% CO2. To detect spontaneous degranulation, effector cells were incubated in the absence of target cells. Effector cells plus PMA (2.5 µg/mL; Biotechne Tocris, Cat. #1201) and ionomycin (0.5 µg/mL; Sigma Aldrich, Cat. #I0634) were the activity positive control. Following co-culture, CD107a expression was analyzed on CD3−CD56+ NK cells by flow cytometry.
Treg suppression assay
PB-derived CD4+CD25+ Tregs and PB-derived CD4+CD25− T effector cells (Teffs) were isolated from fresh PBMCs using the Dynabeads Regulatory CD4+CD25+ T-cell kit (>95% purity tested by flow cytometry; Invitrogen by Life Technologies, Cat. #11363D). Briefly, CD4+ cells were isolated by negative selection using the human CD4 antibody mix. A depletion beads solution was added to remove the non-CD4+ cells. Next, CD25-beads were added to the CD4+ T cells to capture the CD4+CD25+ Tregs and the remaining fraction corresponded to CD4+CD25− Teff cells. All purification steps were performed according to the manufacturer’s instructions. Carboxyfluorescein diacetate succinimidyl ester (CFSE)-labeled autologous PB-derived Teffs (CellTrace CFSE Cell Proliferation Kit, Molecular Probes, by Life Technologies, Cat. #34554) were cultured with PB-derived Tregs at 1:1 ratio in a suppression assay. Cells were cultured (5 × 103 cells/well) in U-bottom 96-well plates with RPMI-1640 medium (GE Healthcare Life Sciences, HyClone Laboratories) supplemented with 2-mmol/L L-glutamine, 100-U/mL penicillin, 100-μg streptomycin, and 10% FBS. Cells were stimulated for 5 days in the presence of Dynabeads Human T-Activator CD3/CD28 in a beads-to-cells ratio of 1:1 (Gibco by Life Technologies, Cat. #11161D) at 37°C in 5% CO2. As a control, Teff cells were cultured in the presence of Dynabeads Human T-Activator CD3/CD28. Tregs suppressive activity was assessed through CFSE-labeled Teffs proliferation by FACS analysis.
Statistical analysis
The statistical distribution of quantitative variables generated by flow cytometry was assessed by the Shapiro-Wilk normality test. All variables were described using the mean and standard deviation (s.d.) or median and range for quantitative variables, according to the distribution of data (i.e., normal or non-normal distribution), and absolute and relative frequency for categorical variables. Outliers were appropriately eliminated. To compare quantitative variables as predictors of clinical response to nivolumab (R and NR), the paired Wilcoxon test or unpaired Mann–Whitney U test for non-normal data was used, whereas paired or unpaired Student t test was used for normal data. For normal data, repeated measures ANOVA with Tukey HSD post hoc test was used to determine treatment effect over time and Bonferroni’s multiple comparison adjustment approach for comparison of expression levels between samples and associations with patient response; for non-normal data, the Friedman test with Dunn’s post hoc test was used to determine treatment effect over time and Bonferroni’s multiple comparison adjustment approach for comparison of expression levels between samples and association patients response. We first evaluated the outcome with each predictor, one at a time, and then considered variables that met a preset cutoff for significance P < 0.05 to run a multivariable model. Second, multivariable logistic regression analyses were performed to identify the independent predictors (continuous) without multicollinearity and to estimate their contribution to the clinical response (dichotomous). The early clinical response prediction performance for each predictor and the best cutoff search were also assessed with receiver operating characteristic curve (ROC) analysis, and the areas under the curve (AUC) were calculated and compared using the DeLong method. Third, the best cutoff was then applied to PFS and OS as endpoints, to estimate the prognostic effect of the predictors. Finally, to generate new biological–clinical hypotheses, exploratory survival analyses were performed to estimate the effect of multiple predictor’s best cutoff categories for predefined subgroups of patients. Statistical analysis was performed using Statistical Package for Social Sciences (version 20, RRID:SCR_002865) and Graphpad Prism (Graphpad Software, Inc., RRID:SCR_002798).
Cross-validation
To improve the results’ robustness, the internal validity of the model was evaluated through 5,000 bootstrap resamples with point estimates and 95% confidence intervals for each cutoff bootstrapped distribution (model evaluation bias, the 0.632+ bootstrap cross-validation technique). For each bootstrap resample, a sample of n = 632 was randomly selected from the original cohort; the probability of inclusion of any participant at each draw was independent of inclusion at previous draws, and therefore, participants could be included in the bootstrap sample multiple times, once, or not at all. The 0.632+ bootstrap cross-validation technique allowed us to evaluate the performance of classifiers and to solve the optimism problem in model evaluation bias. The best cutoff selection was performed by the 0.632+ bootstrap method. Point estimates and 95% confidence intervals for each cutoff bootstrapped distribution with the corresponding AUCs were performed.
Data availability
The data are openly available at https://zenodo.org/records/11394968 and from the corresponding author upon reasonable request.
Results
Patient characteristics and study design
Sixty-nine patients with mRCC were enrolled from April 2016 to June 2022 (11/69 patients according to the nivolumab expanded access program) in the REVOLUTION trial (NCT03891485), a longitudinal, prospective, observational, multicenter study. Patients, who had undergone nivolumab therapy as the second/third/fourth line of therapy for mRCC, were evaluated for peripheral NKs and Tregs, phenotype, and function, at time 0–1–3–6–12 months of treatment (Fig. 1A). Twelve patients were considered not evaluable after T0 evaluation as 8/12 did not return to the academic center and 4/12 died within a month receiving only one nivolumab treatment (early death patients; Fig. 1B). Demographic and clinical/pathological characteristics of patients with mRCC-REV and HDs were reported in Supplementary Tables S2 and S3, respectively. All patients and HDs were Caucasian. Patients and HDs were gender matched. The impact of age, not perfectly matched, was evaluated (Supplementary Fig. S2A–S2J). The study population shows median PFS and OS of 6.0 months (95% CI, 2.8–9.1) and 21.0 months (95% CI, 13.6–28.4), respectively, in accordance with previous results (Fig. 1C; ref. 15). Study–patient representativeness was shown in Supplementary Table S4.
At pretreatment, peripheral samples from patients with mRCC-REV display impaired NKs and a higher frequency of Tregs compared with HDs
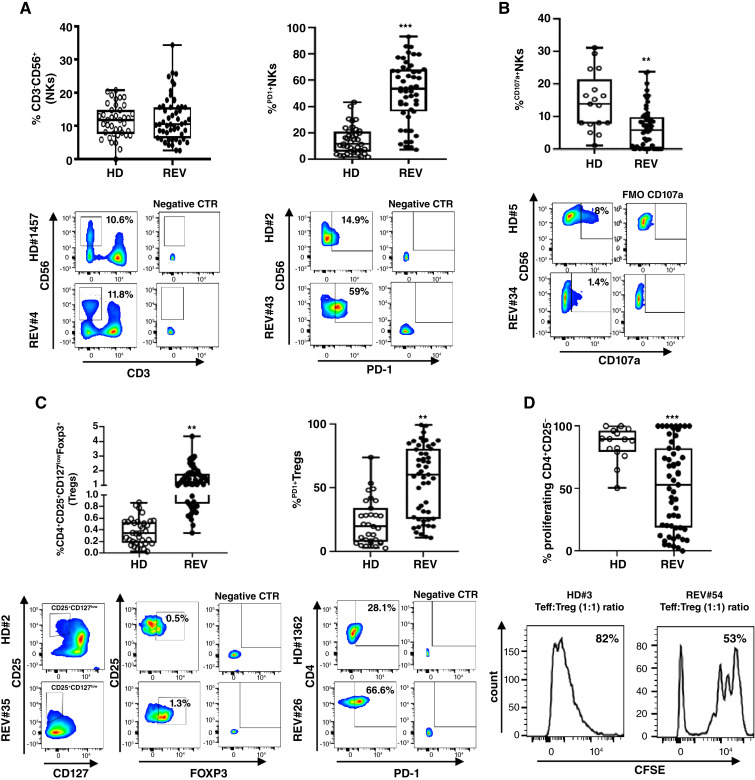
Fifty-seven patients with mRCC-REV were evaluated at the time of the enrolment and compared with 62 HDs. Peripheral immune cell subsets were analyzed through multi-parameter flow cytometry. CD4+T cells, PD1+CD4, CD8+T cells, PD1+CD8, NKs (CD3−CD56+), NKsdim (CD3−CD56dimCD16+), NKsbright (CD3−CD56brightCD16−/+), NKs expressing activating receptors (NKG2D, NKp44, NKp30, and NKp46) or inhibitory receptors (KIR2DL1, KIR2DL2/DL3, and KLRB1 plus PD1), Tregs (CD4+CD25+CD127lowFoxp3+), and Tregs activation markers (CTLA-4, PD1, ICOS, Helios, ENTPD1, and CXCR4) were evaluated (Supplementary Table S5). NKs frequency was unmodified but PD1+NKs were higher as compared with HDs (mean ± s.d.: 50.9% ± 22.9% vs. 16.2% ± 12.4%; P < 0.001, respectively; Fig. 2A). As shown in Fig. 2B, IL2 treated REV-NKs cultured with K562 cells exhibited lower surface CD107a as a measure of NK cytotoxic activity when compared with HDs (mean ± s.d.: 6.4% ± 6.4% vs. 14.4% ± 8.9%; P < 0.01). NKs expressing activating receptor, NKp44+NKs, were significantly lower in patients with REV compared with HDs (mean ± s.d.: 3% ± 3.5% vs. 14.6% ± 5.5%; P < 0.001), whereas NKp46+NKs and NKp30+NKs frequencies were higher (mean ± s.d.: 76.9% ± 37.2% vs. 39.9% ± 23.9%; P < 0.001 and 68.9% ± 21% vs. 53.6% ± 16.2%; P < 0.01; Supplementary Fig. S3A). In terms of inhibitory receptors, the frequency of KIR2DL1+NKs and KIR2DL2/DL3+NKs were higher as compared with HDs (mean ± s.d.: 10.4% ± 9% vs. 4.6% ± 4.8%; P < 0.001; 34.1% ± 14.5% vs. 27.7% ± 10.9%; P < 0.05; Supplementary Fig. S3B). The subset of NKs co-expressing PD1 and KIR2DL1 or PD1 and KIR2DL2/DL3 was significantly higher in patients REV as compared with HDs (mean ± s.d.: 8.6% ± 9.9% vs. 3.1% ± 2.9%; P < 0.01; 16.2% ± 11.2% vs. 10.3% ± 6.5%; P < 0.01, respectively; Supplementary Fig. S4) in accordance with previous results (37, 38). A high frequency of peripheral Tregs (CD4+CD25+CD127lowFoxp3+) and PD1+Tregs was observed in patients with REV vs. HD (mean ± s.d.: 1.4% ± 0.8% vs. 0.4% ± 0.2%; P < 0.01; 55.7% ± 27.6% vs. 23% ± 17.8%; P < 0.01; Fig. 2C). Tregs from patients with REV were more suppressive to T effectors than HDs in co-culture assay (mean ± s.d.: 51.7% ± 34.9% vs. 85.4% ± 13.7%; P < 0.001 Teff proliferation; Fig. 2D). Moreover, immunosuppressive Helios+Tregs and ENTPD1+Tregs were significantly higher in patients with REV compared with HDs (mean ± s.d.: 49.2% ± 33.4% vs. 19.5% ± 7.6%; P < 0.05; 50.2% ± 27.8% vs. 33.9% ± 26.4%; P < 0.01; Supplementary Fig. S3C). Finally, PD1+CD4 and PD1+CD8 were significantly higher in patients with REV as compared with HDs (mean ± s.d.: 49.7% ± 33.0% vs. 29.1% ± 18.0%; P < 0.05; 38.0% ± 27.2% vs. 18.7% ± 8.9%; P < 0.01; Supplementary Fig. S3D). Altogether, these data reveal that the PB of patients with REV displayed pretreatment defective NKs and a higher frequency of Tregs endowed with increased suppressive functions as compared with HDs.
Figure 2.
Pretreatment NKs and Tregs in patients with mRCC-REV. A, Upper: % NKs and % PD1+NKs in HD and REV groups. % NKs: HD (n = 39) vs. REV (n = 54), P = NS; % PD1+NKs: HD (n = 37) vs. REV (n = 51), P < 0.001. Lower: Representative comparison between HD#1457 and REV#4, HD#2 and REV#43. B, Upper: % CD107a+NKs in HD (n = 17) and REV (n = 56) groups, P < 0.05. Lower: Representative comparison between HD#5 and REV#34. C, Upper: % Tregs in HD (n = 34) and REV (n = 55) groups, P < 0.01. % PD1+Tregs: HD (n = 31) vs. REV (n = 51), P < 0.01. Lower: representative comparison between HD#2 and REV#35, HD#1362 and REV#26. D, Upper: % CFSE-Teff proliferation/Tregs dependent in HD (n = 15) vs. REV (n = 55) groups, P < 0.001. Lower: Representative comparison between HD#3 and REV#54. Box plot showing the data distribution across groups with minimum, median, and maximum values. Two-tailed test comparison of differences between two independent groups (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Peripheral NKs and Tregs did not change significantly during nivolumab treatment in patients with REV
NKs and Tregs, phenotype, and function, were longitudinally evaluated in PB; 45-paired samples were analyzed at time 0–1–3 months (Supplementary Fig. S5; Supplementary Table S6), showing that the total frequency of NKs does not significantly change over time. NKp30+NKs significantly decreased post 1 and 3 months of treatment, whereas KIR2DL1+NKs significantly increased after 1 month of treatment (Supplementary Fig. S5A). CTLA-4+Tregs modestly but significantly decreased after 3 months of therapy (Supplementary Fig. S5C).
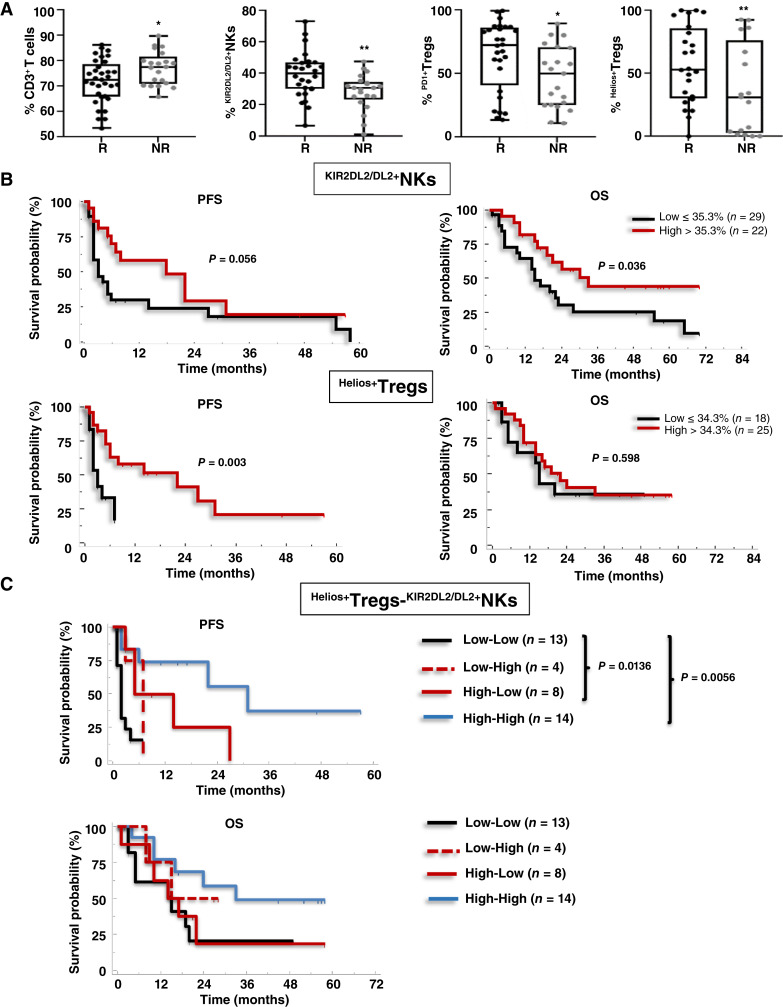
Pretreatment peripheral low CD3+, high KIR2DL2/DL3+NKs, high Helios+Tregs, and PD1+Tregs profile identifies patients with nivolumab response
Based on 3 months of clinical response, patients were classified as Rs (PR, SD ≥6 months; 33/55) and NRs (SD <6 months, PD, D; 22/55; Supplementary Fig. S6). At pretreatment, R patients presented significantly lower CD3+ (mean ± s.d.: 71.9% ± 8.6% vs. 77% ± 6.5%; P < 0.05), higher KIR2DL2/DL3+NKs (mean ± s.d.: 39.1% ± 14.4% vs. 28.3% ± 11.4%; P < 0.001), higher PD1+Tregs (mean ± s.d.: 64.4% ± 27.5% vs. 47.3% ± 24.4%; P < 0.05) and Helios+Tregs (mean ± s.d.: 60.4% ± 30.2% vs. 32.2% ± 33.2%; P < 0.01; Fig. 3A; Supplementary Table S7). This suggests that R patients display at baseline a less efficient immune peripheral phenotype, with defective NKs and higher Tregs, than the group as a whole. We performed the same analysis again, including the four patients who died within 1 month of enrolment as NR; the results were identical (Supplementary Table S8). We also noted higher pretreatment frequencies of KIR2DL2/DL3+NK, PD1+Tregs, and Helios+Tregs in patients who reached 12 months of treatment (LR, Long Responders; Supplementary Fig. S7). These immune populations were evaluated as predictor variables in a multivariate test predicting 3 months of clinical response. Collinearity diagnostics showed a high correlation between PD1+Tregs and Helios+Tregs, so PD1+Tregs were excluded from the analysis. The logistic regression methods identified CD3+ (P = 0.029; OR: 1.228; 95% CI, 1.022–1.475), KIR2DL2/DL3+NKs (P = 0.047; OR: 0.929; 95% CI, 0.863–0.999) and Helios+Tregs (P = 0.025; OR: 0.963; 95% CI, 0.932–0.995) each as independent predictors of nivolumab responsiveness (Table 1). Through ROC curve and 0.632+ cross-validation bootstrap methods, R from NR cutoff values were identified. ROC curve relative to pretreatment KIR2DL2/DL3+NKs and Helios+Tregs as nivolumab predictive markers displayed statistically significant results for both AUCs estimated by 0.632+ internal cross-validation approach equal to 0.724 (P < 0.001) for KIR2DL2/DL3+NKs and to 0.741 (P < 0.001) for Helios+Tregs. The KIR2DL2/DL3+NKs optimally selected cutoff point of 35.3% (95% CI, 25.8–43.0) resulted in 80.95% sensitivity, 60.71% specificity with a positive predictive value (PPV) 81% (probability of correctly identifying a nivolumab R patient when the value of KIR2DL2/DL3+NKs is >35.3%) and negative predictive value (NPV) 60.7% (probability of correctly identifying a nivolumab NR patient when the value of KIR2DL2/DL3+NKs is ≤35%). The Helios+Tregs optimally selected cutoff point of 34.3% (95% CI, 24.5–45.2) resulted in 70.6% sensitivity, 75.0% specificity, with a PPV of 78.3% and NPV of 66.7% (Supplementary Fig. S8; Supplementary Table S9).
Figure 3.
Pretreatment low CD3+, high KIR2DL2/DL3+NKs, PD1+Tregs and Helios+Tregs predict nivolumab response. A, Frequencies of CD3+ (as a percentage of lymphocytes) and KIR2DL2/DL3+NKs, PD1+Tregs, and Helios+Tregs in R vs. NR patients. CD3+: (33 R vs. 31 NR; P < 0.05); KIR2DL2/DL3+NKs: (28 R vs. 21 NR; P < 0.01); PD1+Tregs: (28 vs. 21 NR; P < 0.05); Helios+Tregs: (24 R vs. 17 NR; P < 0.01). Box plot showing the data distribution across groups with minimum, median, and maximum values. Two-tailed test comparison of differences between two independent groups (*, P < 0.05; **, P < 0.01; ***, P < 0.001). B, Kaplan–Meier survival curves for PFS and OS of patients with mRCC-REV, stratified according to optimal cutoff identified by ROC analyses; C, Kaplan–Meier plots for PFS according to combination of Helios+Tregs-KIR2DL2/DL3+NKs values. Low–Low (black line); Low–High (dotted red line); High–Low (red line); High–High (blue line). P-values by log-rank test.
Table 1.
Logistic regression predicting likelihood of nivolumab response.
| (%) | B | SE | Wald | df | P | Odds ratio | 95% CI for odds ratio | |
|---|---|---|---|---|---|---|---|---|
| Lower | Upper | |||||||
| T = 0 | ||||||||
| CD3+ | 0.205 | 0.094 | 4.790 | 1.000 | 0.029 | 1.228 | 1.022 | 1.475 |
| KIR2DL2/DL3+NKs | −0.074 | 0.037 | 3.932 | 1.000 | 0.047 | 0.929 | 0.863 | 0.999 |
| Helios+Tregs | −0.038 | 0.017 | 5.024 | 1.000 | 0.025 | 0.963 | 0.932 | 0.995 |
| Constant | −11.610 | 6.212 | 3.493 | 1.000 | 0.062 | 0.000 | ||
| T = 1 month of treatment | ||||||||
| CD3+ | 0.144 | 0.064 | 5.119 | 1.000 | 0.024 | 1.155 | 1.019 | 1.309 |
| KIR2DL2/DL3+NKs | −0.066 | 0.032 | 4.185 | 1.000 | 0.041 | 0.936 | 0.878 | 0.997 |
| ICOS+Tregs | −0.033 | 0.024 | 1.865 | 1.000 | 0.172 | 0.968 | 0.923 | 1.014 |
| Constant | −8.031 | 4.726 | 2.888 | 1.000 | 0.089 | 0.000 | ||
Multivariate logistic regression method enter; variable or predictor at basal level: % CD3+, % KIR2DL2/DL3+NKs and % Helios+Tregs; variable or predictor after 1 month of treatment: % CD3+, % KIR2DL2/DL3+NKs and % ICOS+Tregs; P value significance < 0.05; B: the estimated coefficient; SE: standard error around the co‐efficient. Wald chi-square statistics; df: degree of freedom for Wald chi-square statistics; Exp (B): exponentiation of B coefficient is an OR predicted change in odds for a unit increase in the predictor; CI: 95% confidence interval for the odds ratio with its upper and lower limits.
Pretreatment peripheral KIR2DL2/DL3+NKs and Helios+Tregs predict survival in patients with REV
To evaluate the prognostic value of pretreatment KIR2DL2/DL3+NKs and Helios+Tregs, Kaplan–Meier curves were developed based on the above-defined optimal cutoff value. Patients with REV with pretreatment KIR2DL2/DL3+NKs >35.3% displayed significantly longer OS (median 33.0 vs. 15.0 months; P = 0.036). In contrast, patients with REV that at pretreatment presented Helios+Tregs >34.3% displayed longer PFS (median 22.0 vs. 3.0 months; P = 0.003; Fig. 3B). The evaluation of Helios+Tregs and KIR2DL2/DL3+NKs were combined in four groups: 1. Low–Low, 2. Low–High, 3. High–Low, and 4. High–High. As shown in Fig. 3C, patients with Helios+Tregs-KIR2DL2/DL3+NKs High–High and High–Low showed a longer PFS compared with Low–Low patients (median 31 vs. 2 months; P = 0.0056 and 9.5 vs. 2 months; P = 0.0136, respectively). DCR was also evaluated for 37 patients with REV expressing both KIR2DL2/DL3+NKs and Helios+Tregs with clinical response available. 13/37 (35.1%) express high Helios+Tregs >34.3%-high KIR2DL2/DL3+NKs >35.3% (High–High) at pretreatment. In this group, the DCR (PR + SD/PR + SD + PD × 100) was 77% (10/13 were R patients). Conversely, in 13/37 (35.1%) that express low Helios+Tregs <34.3%–low KIR2DL2/DL3+NKs <35.3% (Low–Low) the DCR was 15% (2/13 were R patients), (Supplementary Fig. S9; Supplementary Table S10). These results suggest that response is most likely to occur in patients that at pretreatment display a specific NK subgroup expressing KIR2DL2DL3 receptors and Tregs expressing Helios and thus the elevated pretreatment KIR2DL2/DL3+NKs and Helios+Tregs represent suitable biomarkers for PFS/OS in nivolumab-treated patients with mRCC.
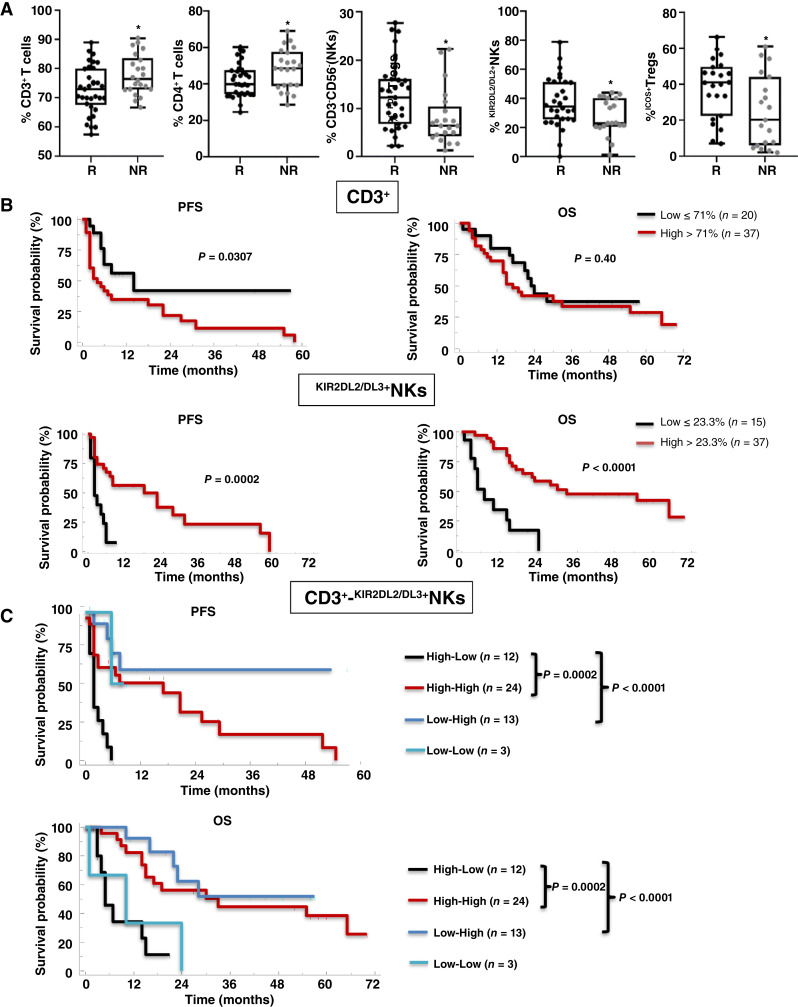
After 1 month of nivolumab, CD3+ and KIR2DL2/DL3+NK frequencies predict response and correlate with survival in patients with REV
After confirming the importance of pretreatment rates of NK and Tregs, we explored the predictive role of peripheral NK and Treg phenotypes after 1 month of nivolumab treatment (T1). Evaluating the PB analysis after 1 month of treatment with the response status of patients, R patients displayed lower CD3+ (mean ± s.d.: 72.8% ± 8.1% vs. 77.9% ± 6.9%; P < 0.05) and CD4+ (mean ± s.d.: 42.2% ± 9.1% vs. 48.3% ± 11.1%; P < 0.05), and higher NKs (mean ± s.d.: 12.8% ± 7% vs. 8.2% ± 5.9%; P < 0.05), KIR2DL2/DL3+NKs (mean ± s.d.: 37.5% ± 17.3% vs. 27.6% ± 12.5%; P < 0.05) and ICOS+Tregs (mean ± s.d.: 38.1% ± 16.3% vs. 25.1% ± 19.9%; P < 0.05), than NR (Fig. 4A; Supplementary Table S11). When the immune cell subsets were stated as predictor variables in a multivariate test, collinearity diagnostics showed a high correlation between the frequencies of CD3+, CD4+, and NKs. Thus, CD4+ and total NKs were excluded by multivariate analysis. Through logistic regression methods, CD3+ (P = 0.024; OR: 1.155; 95% CI, 1.019–1.309) and KIR2DL2/DL3+NKs (P = 0.041; OR: 0.936; 95% CI, 0.878–0.997) were independent predictors of nivolumab responsiveness (Table 1). About CD3+ and KIR2DL2/DL3+NKs as predictive biomarkers of nivolumab response at T1, the ROC curve displayed statistically significant results for both AUCs estimated by 0.632+ internal cross-validation approach equal to 0.672 (P < 0.001) for CD3+ and equal to 0.677 (P < 0.001) for KIR2DL2/DL3+NKs. The CD3+ optimally selected cutoff point of 71% (95% CI, 59.6–83.2) resulted in 86.4% sensitivity, and 48.5% specificity, with a PPV of 84% (probability of correctly identifying a nivolumab R patient when the value of CD3+ is ≤71%) and a NPV of 52.7% (probability of correctly identifying a nivolumab NR patient when the value of CD3+ is >71%). The KIR2DL2/DL3+NKs optimally selected cutoff point of 23.3% (95% CI, 14.4–31.5) resulted in 55.5% sensitivity, 86.7% specificity, with a PPV of 74.3 and NPV of 73.3% (Supplementary Fig. S10; Supplementary Table S12). Here, the association of low frequencies of CD3+ cells and high KIR2DL2/DL3+NKs with response may reflect the impact of nivolumab on the entire T-cell compartment and confirm the role of KIR2DL2/DL3 NKs as discussed below.
Figure 4.
One-month nivolumab- CD3+ and KIR2DL2/DL3+NKs predict response. A, Frequencies of CD3+, CD4+, NKs and KIR2DL2/DL3+NKs, and ICOS+Tregs in R vs. NR patients. CD3+ T cells: 33 R vs. 22 NR, P < 0.05; CD4+ T cells: 33 R vs. 22 NR, P < 0.05; NKs: 31 R vs. 21 NR, P < 0.05; KIR2DL2/DL3+NKs: 30 R vs. 21 NR, P < 0.05; ICOS+Tregs: 23 R vs. 19 NR, P < 0.05. Two-tailed test comparison of differences between two independent groups (P < 0.05). Box plot showing the data distribution across groups with minimum, median, and maximum values. Two-tailed test comparison of differences between two independent groups (*, P < 0.05; **, P < 0.01; ***, P < 0.001). B, Kaplan–Meier survival curves for PFS and OS of patients with mRCC-REV stratified according to optimal cutoff identified by ROC analyses. C, Kaplan–Meier plots showing PFS and OS of patients stratified according to a combination of CD3+-KIR2DL2/DL3+NKs. High–Low (black line); High–High (red line); Low–High (blue line); Low–Low (light blue line). P-values by long-rank test.
After 1 month of nivolumab, low frequency of CD3+ (≤71%) significantly correlated with longer PFS (median 20 vs. 4 months; P = 0.0307) but not with OS (median 23 vs. 17 months; P = 0.40), whereas higher frequency of KIR2DL2/DL3+NKs (>23.3%) associated with both PFS (median 22 vs. 2 months; P = 0.0002) and OS (median 33 vs. 7 months; P < 0.0001; Fig. 4B). Extending these remarkable differences, combining CD3+ and KIR2DL2/DL3+NKs values showed that patients with low CD3+(≤71%) and high KIR2DL2/DL3+NKs (>23.3%), or high CD3+ and high KIR2DL2/DL3+NKs after 1 month of nivolumab have longer PFS and OS as compared with high CD3+ and low KIR2DL2/DL3+NKs (Low–High vs. High–Low, PFS: median survival not reached vs. 2 months; P < 0.0001; OS: median not reached vs. 5 months; P < 0.0001; High–High vs. High–Low, PFS: median survival 18 vs 2 months; P = 0.0002; OS: median 33 vs. 5 months; P = 0.002, respectively; Fig. 4C). In Fig. 5 a recapitulating cartoon is reported.
Figure 5.
Schematic representative. A, Nivolumab-treated patients with mRCC are evaluated for peripheral NKs and Tregs, phenotype, and function. Clinical evaluation at 3 months identifies patients R and NR. B, Pretreatment (T0) evaluation of NKs and Tregs identified R patients (high KIR2DL2/DL3+NKs and high Helios+Tregs) with improved prognosis. C, One-month post–nivolumab treatment evaluation of NKs and Tregs identified R patients (low CD3+ and high KIR2DL2/DL3+NKs) with improved prognosis.
Discussion
REVOLUTION is a peripheral, longitudinal study aimed at identifying biomarkers predicting nivolumab response in patients with mRCC. NKs and Tregs, phenotype and function, were analyzed in 57 patients with mRCC-REV at pretreatment and during nivolumab treatment. At pretreatment, patients with mRCC-REV displayed a high percentage of PD1+CD4+/CD8+, defective NKs, and increased suppressive Tregs as compared with HDs. In particular, patients with mRCC were characterized by a high frequency of NKs expressing inhibitory receptors (KIR2DL1, KIR2DL2/DL3, and PD1) and Tregs expressing activation markers (PD1, Helios, and ENTPD1). Clinical evaluation of nivolumab response at 3 months stratified patients as R and NR. At the pretreatment evaluation, R patients exhibited higher frequencies of KIR2DL2/DL3+NKs and Helios+Tregs. According to the defined cutoff, KIR2DL2/DL3+NKs >35.5% and Helios+Tregs >34.3% predicted nivolumab sensitivity and prognosis. Therefore, we suggest that the peripheral pretreatment level of KIR2DL2/DL3+NKs and Helios+Tregs represents an easy and noninvasive biomarker of response to nivolumab in patients with mRCC, alone and even more accurately in combination. Additionally, we explored the predictive role of peripheral NKs and Tregs after 1 month of nivolumab treatment. The frequency of lower CD3+ and higher KIR2DL2/DL3+NKs correlated with clinical benefit and survival in patients with REV.
Our observation of a higher frequency of NK expressing the inhibitory receptor KIR2DL2/DL3 associated with nivolumab responsiveness and longer OS initially appears counterintuitive. NK cells exert selective cytotoxicity against tumor cells largely based on MHC class I-specific inhibitory receptors KIRs and CD94-NKG2A (28). Tumor MHC-I heterogeneity affects biological aggressiveness, metastatic potential, and immunotherapy sensitivity (39). MHC-I deficient tumor cells derive from T-cell immune selection in primary heterogeneous tumors (40) and cancer cells lacking the MHC-I molecule will interrupt MHC-I-KIR interaction (“missing-self recognition”). KIR2DL1-3 binds to the ligand HLA-C, expressed by tumor cells and other nucleated cells, to prevent the activation of NK cells, thereby regulating the cytotoxicity of NK cells and inducing self-tolerance (41, 42). Interestingly patients with triple-negative breast cancer carrying KIR2DL2+/HLA-C1+ displayed improved clinical response to pegylated–IL2 plus nivolumab (43). Conversely, in patients with metastatic solid tumors, no associations were identified between KIR-ligand present/missing status and prognosis (e.g., PIVOT-02 trial evaluating pegylated –IL2 with nivolumab; refs. 43, 44). The acquisition of KIR surface expression by NK cells derives from the process called “licensing”. During maturation, NK cells modulate their activation response and increase functional activity (45). Licensing derives from the recognition of the corresponding HLA-I complex on healthy cells by an inhibitory KIR and the subsequent transduction of the inhibitory signal through the inner domain of the receptor (46–48). Patients with the KIR-ligands present genotype (HLA-C1) may have more “licensed” (namely more potent) NKs that, in this setting, may not be inhibited because of the low level of HLA on their tumors and thus be associated with a greater antitumor effect (43). As previously reported, 84% of patients with mRCC (88/107) showed HLA-C1, the ligand for KIR2DL2/DL3 (49), and in 20 primary RCC tumor samples 45% (9/20) partial/or complete loss of HLA-I molecules was reported (50). Preliminary evidence showed that nivolumab R patients displayed tumoral low HLA-I (-A,-B,-C) as compared with NR patients, and HLA-I expression inversely correlated to pretreatment peripheral %KIR2DL2/DL3+NKs (Supplementary Fig. S11). Thus, we can speculate that NKs of R patients are more “licensed” and PD1 inhibition in patients with tumor cells with low or no HLA-I expression results in NK activation.
High pretreatment frequency of active peripheral Tregs (expressing PD1, Helios, and/or ENTPD1) was previously described in primary and patients with mRCC (51–53). The effects of anti-PD1 on Tregs are not completely clear. In patients with hyperprogressed gastric cancer, nivolumab increased the proliferation of tumor-infiltrating Tregs and potentiated the in vitro suppressive activity of peripheral Tregs (54). On the contrary, ex vivo CXCR4 antagonist/nivolumab treatment of peripheral Tregs from patients with primary RCC decreased Tregs suppressive function while increasing IFN-γ secretion (53). Consistent with that, in vitro blockade of PD1 increased the proliferation of peripheral Tregs and pSTAT3 expression while reducing Treg-suppressive function in patients with melanoma (55). In this study, we observed pretreatment higher percent of PD1+Tregs and Helios+Tregs in nivolumab R patients. As was previously reported, circulating PD1+Tregs potentially predict anti-PD1 response in patients with melanoma. In particular, patients with melanoma responding to nivolumab displayed high pretreatment PD1+Tregs, which may be modulated by the PD1 blockers (56). Consistent with this, our data show that pretreatment Helios+Tregs predicts nivolumab response. The meaning of Helios+Treg cells is not entirely clear, with conflicting results that are dependent on context and Treg evaluation as distinct functional subpopulations (57, 58), FOXP3 stability (59), epigenetic changes (60, 61), and function as CFSE assay proliferation (62, 63). In cancer, low or high Helios+Tregs were linked with poor prognosis (57, 64, 65). High CTLA-4, CD39, TIGIT, and PD1 were associated with highly immunosuppressive Helios+Tregs in NSCLC and colorectal cancer (66, 67). Herein, Helios+Tregs showed a moderate correlation with PD1+Tregs in R patients (Kendall’s coefficient 0.396; P < 0.015) but not in the NR group (Kendall’s coefficient 0.125; P < 0.48; Supplementary Fig. S12) suggesting that in R patients high PD1+Tregs and Helios+Tregs could identify a population more sensitive to anti-PD1 therapy, although Helios seems a marker, more than a driver, of human Treg stability (58). To support it, although not significant, pretreatment Tregs isolated from R patients were less able to inhibit T effector cell proliferation as compared with NR Tregs, allowing the speculation that PD1+Tregs and Helios+Tregs may not necessarily reflect suppressive Tregs (58, 61, 68). Of note, after 1 month of nivolumab, a trend in the decrease of PD1+Tregs and Helios+Tregs was observed in R patients (Supplementary Fig. S13), suggesting that anti-PD1 could exert an antitumor effect reducing active Tregs. Preliminary evidence showed that after 1 month of nivolumab, only Tregs from R patients were inhibited by the newly developed CXCR4 antagonist R54 (69), suggesting that CXCR4 may play a crucial role in inhibiting Tregs function in mRCC as in primary RCC (70). Moreover, patients with mRCC with high pretreatment peripheral PD1+Tregs and Helios+Tregs, had a better response to anti-PD1 therapy suggesting PD1 blockade may attenuate the immune-suppressive function of Tregs.
Taken together, these results suggest that pretreatment or early on-treatment (1 month) evaluation of KIR2DL2/DL3+NKs and Helios+Tregs or CD3+ and KIR2DL2/DL3+NKs, respectively, represents early, feasible, predictive and prognostic biomarkers in nivolumab-treated patients with mRCC and may help in categorizing patients who will benefit from nivolumab. Collection of these biomarkers will be evaluated in an ongoing study of first-line ICI–tyrosine kinase inhibitor versus ICI–ICI in patients with mRCC.
Supplementary Material
Supplementary Table S1. Antibodies used for flow cytometry Supplementary Table S2. mRCC-REV patients characteristics Supplementary Table S3. HDs characteristics Supplementary Table S4. Representativeness of Study Participants Supplementary Table S5. Univariate analysis of peripheral immune cell subsets between HD and mRCC-REV groups Supplementary Table S6. Longitudinal evaluation of peripheral immune cell subsets in mRCC-REV patients Supplementary Table S7. Differential distribution of peripheral immune cell subsets between R and NR groups (T0) in mRCC-REV patients Supplementary Table S8. Univariate analysis of peripheral immune cell subsets between R and NR groups including early death patients (T0) in mRCC-REV patients Supplementary Table S9. Cut-off value prediction with ROC curve for continuous variable at basal level and response to nivolumab after 3 months as classification variable Supplementary Table S10. Association between the KIR2DL2/DL3+NKs/Helios+Tregs values and response to nivolumab treatment Supplementary Table S11. Differential distribution of peripheral immune cell subsets between R and NR groups (T1) in mRCC-REV patients Supplementary Table S12. Cut-off value prediction with ROC curve for continuous variable at T1 and response to nivolumab after 3 months as classification variable
Supplementary Figure S1. Gating strategy for identification of Tregs and NKs in peripheral blood.
Supplementary Figure S2. Correlation between age and pretreatment NKs and Tregs immune subsets cells in mRCC-REV patients and HDs.
Supplementary Figure S3. Pretreatment NKs and Tregs in mRCC-REV patients
Supplementary Figure S4. Pretreatment NKs co-expression of PD-1 and KIR2DL1or KIR2DL2/DL3 in mRCC-REV patients.
Supplementary Figure S5. Nivolumab did not significantly affect peripheral NKs and Tregs
Supplementary Figure S6. Swimmer’s plot of nivolumab-treated mRCC patients.
Supplementary Figure S7. Pretreatment NKs and Tregs subpopulations in mRCC-REV Long Responder (LR) patients vs Not-Responders (NR).
Supplementary Figure S8. .632+ cross validation bootstrap ROC curves and the optimum pretreatment cut-off values of KIR2DL2/DL3+NKs and Helios+Tregs to determine the outcome of REV patients
Supplementary Figure S9. Association between the KIR2DL2/DL3+NKs/Helios+Tregs values and response to nivolumab treatment.
Supplementary Figure S10. .632+ cross validation bootstrap ROC curves and the optimum T1 cut-off values of CD3+ and KIR2DL2/DL3+NKs to determine the outcome of REV patients
Supplementary Figure S11. HLA class I in primary RCC from REV patients
Supplementary Figure S12. Correlation between pretreatment of PD-1+Tregs and Helios+Tregs in Responder and Not-Responder patients.
Supplementary Figure S13. T0-T1 values (after one month of nivolumab) between PD-1+ Tregs and Helios+Tregs in Responder and Not Responder patients
Acknowledgments
This work was supported by the following grants: 1. “Prediction of nivolumab action in patients with metastatic renal cancer: Treg function, tumoral access, and NK interactions as predictive biomarkers of immunotherapy (REVOLUTION)”-ERA-NET/TRANSCAN REVOLUTION (TRS-2016-0000034); 2. “Approccio integrato per la caratterizzazione prognostico-predittiva del microambiente tumorale nel cancro renale” Ministero della Salute (m2/6-c, n.659, August 20, 2018); 3. “New generation of CXCR4 antagonist: peptide R54 from biomolecular mechanism to first in man clinical study” funded by AIRC (ID_24746, n.1018, October 08, 2021). S. Scala is the recipient of the above-cited grants.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors’ Disclosures
H.J. Boyle reports non-financial support from BMS, Pfizer, Ipsen, and Merck outside the submitted work. J. Carles reports personal fees from Astellas Pharma, AstraZeneca, Bayer, Exelixis, Johnson & Johnson, Novartis (AAA), Pfizer, Sanofi, BMS, Ipsen, and Roche and grants from AB Science, Aragon Pharmaceuticals, Arog Pharmaceuticals, Astellas Pharma, AstraZeneca AB, Aveo Pharmaceuticals, Bayer AG, Blueprint Medicines Corporation, BN Immunotherapeutics, Boehringer Ingelheim España SA, Bristol-Myers Squibb International Corporation (BMS), Clovis Oncology, Cougar Biotechnology, Deciphera Pharmaceuticals, Exelixis, F. Hoffmann-La Roche, Genentech, GlaxoSmithKline SA, Incyte Corporation, Janssen-Cilag International NV, Laboratoires Leurquin Mediolanum SAS, Lilly SA, Medimmune, Millennium Pharmaceuticals, Nanobiotix SA, Novartis Farmacéutica SA, Pfizer SLU, Puma Biotechnology, Sanofi-Aventis SA, SFJ Pharma LTD II, and Teva Pharma SLU outside the submitted work. S. Pignata reports grants and personal fees from MSD, GSK, Roche, and AstraZeneca and personal fees from Novartis outside the submitted work. No conflicts of interest were disclosed by the other authors.
Authors’ Contributions
S. Santagata: Conceptualization, data curation, formal analysis, methodology, writing–original draft, writing–review and editing. A.M. Trotta: Conceptualization, data curation, formal analysis, methodology, writing–original draft, writing–review and editing. C. D’Alterio: Conceptualization, data curation, formal analysis, methodology, writing–original draft, writing–review and editing. M. Napolitano: Conceptualization, data curation, formal analysis, methodology, writing–review and editing. G. Rea: Conceptualization, data curation, formal analysis, methodology, writing–review and editing. M. Di Napoli: Conceptualization, resources, data curation, writing–review and editing. L. Portella: Data curation, methodology, writing–review and editing. C. Ieranò: Data curation, formal analysis, methodology. G. Guardascione: Data curation, formal analysis, methodology. E. Coppola: Resources, data curation, formal analysis. C. Caux: Conceptualization, resources, writing–original draft, writing–review and editing. B. Dubois: Conceptualization, resources, writing–original draft, writing–review and editing. H.J. Boyle: Conceptualization, resources, writing–original draft, writing–review and editing. J. Carles: Conceptualization, resources, writing–original draft, writing–review and editing. S. Rossetti: Conceptualization, resources, data curation, writing–review and editing. R. Azzaro: Conceptualization, resources. F. Feroce: Conceptualization, resources, formal analysis. S. Perdonà: Conceptualization, resources, writing–original draft, writing–review and editing. M. Fordellone: Data curation, methodology, writing–original draft, writing–review and editing. A.M. Bello: Data curation, formal analysis, methodology. D. Califano: Conceptualization, data curation, writing–original draft, writing–review and editing. P. Chiodini: Conceptualization, data curation, formal analysis, methodology, writing–original draft, writing–review and editing. S. Pignata: Conceptualization, resources, data curation, formal analysis, supervision, writing–original draft, writing–review and editing. S. Scala: Conceptualization, data curation, formal analysis, supervision, funding acquisition, validation, writing–original draft, writing–review and editing.
References
- 1. Ferlay J, Colombet M, Soerjomataram I, Mathers C, Parkin DM, Piñeros M, et al. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer 2019;144:1941–53. [DOI] [PubMed] [Google Scholar]
- 2. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021;71:209–49. [DOI] [PubMed] [Google Scholar]
- 3. Hsieh JJ, Purdue MP, Signoretti S, Swanton C, Albiges L, Schmidinger M, et al. Renal cell carcinoma. Nat Rev Dis Primers 2017;3:17009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gnarra JR, Tory K, Weng Y, Schmidt L, Wei MH, Li H, et al. Mutations of the VHL tumour suppressor gene in renal carcinoma. Nat Genet 1994;7:85–90. [DOI] [PubMed] [Google Scholar]
- 5.The Cancer Genome Atlas Research Network; Morgan M, Gunaratne PH, Wheeler DA, Gibbs RA, Gordon Robertson A, et al. The Cancer Genome Atlas Research Network . Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 2013;499:43–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Athanazio DA, Amorim LS, da Cunha IW, Leite KRM, da Paz AR, de Paula Xavier Gomes R, et al. Classification of renal cell tumors – current concepts and use of ancillary tests: recommendations of the Brazilian Society of Pathology. Surg Exp Pathol 2021;4:4. [Google Scholar]
- 7. Roberto M, Botticelli A, Panebianco M, Aschelter AM, Gelibter A, Ciccarese C, et al. Metastatic renal cell carcinoma management: from molecular mechanism to clinical practice. Front Oncol 2021;11:657639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tran J, Ornstein MC. Clinical review on the management of metastatic renal cell carcinoma. JCO Oncol Pract 2022;18:187–96. [DOI] [PubMed] [Google Scholar]
- 9. Motzer RJ, Tannir NM, McDermott DF, Arén Frontera O, Melichar B, Choueiri TK, et al. Nivolumab plus ipilimumab versus sunitinib in advanced renal-cell carcinoma. N Engl J Med 2018;378:1277–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Escudier B. Combination therapy as first-line treatment in metastatic renal-cell carcinoma. N Engl J Med 2019;380:1176–8. [DOI] [PubMed] [Google Scholar]
- 11. McDermott DF, Huseni MA, Atkins MB, Motzer RJ, Rini BI, Escudier B, et al. Clinical activity and molecular correlates of response to atezolizumab alone or in combination with bevacizumab versus sunitinib in renal cell carcinoma. Nat Med 2018;24:749–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Braun DA, Hou Y, Bakouny Z, Ficial M, Sant’ Angelo M, Forman J, et al. Interplay of somatic alterations and immune infiltration modulates response to PD-1 blockade in advanced clear cell renal cell carcinoma. Nat Med 2020;26:909–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Janiszewska AD, Poletajew S, Wasiutyński A. Spontaneous regression of renal cell carcinoma. Contemp Oncol (Pozn) 2013;17:123–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jian Y, Yang K, Sun X, Zhao J, Huang K, Aldanakh A, et al. Current advance of immune evasion mechanisms and emerging immunotherapies in renal cell carcinoma. Front Immunol 2021;12:639636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Motzer RJ, Escudier B, McDermott DF, George S, Hammers HJ, Srinivas S, et al. Nivolumab versus everolimus in advanced renal-cell carcinoma. N Engl J Med 2015;373:1803–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xu W, Atkins MB, McDermott DF. Checkpoint inhibitor immunotherapy in kidney cancer. Nat Rev Urol 2020;17:137–50. [DOI] [PubMed] [Google Scholar]
- 17. Monjaras-Avila CU, Lorenzo-Leal AC, Luque-Badillo AC, D’Costa N, Chavez-Muñoz C, Bach H. The tumor immune microenvironment in clear cell renal cell carcinoma. Int J Mol Sci 2023;24:7946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Au L, Hatipoglu E, Robert de Massy M, Litchfield K, Beattie G, Rowan A, et al. Determinants of anti-PD-1 response and resistance in clear cell renal cell carcinoma. Cancer Cell 2021;39:1497–518.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kumagai S, Togashi Y, Kamada T, Sugiyama E, Nishinakamura H, Takeuchi Y, et al. The PD-1 expression balance between effector and regulatory T cells predicts the clinical efficacy of PD-1 blockade therapies. Nat Immunol 2020;21:1346–58. [DOI] [PubMed] [Google Scholar]
- 20. Braun DA, Ishii Y, Walsh AM, Van Allen EM, Wu CJ, Shukla SA, et al. Clinical validation of PBRM1 alterations as a marker of immune checkpoint inhibitor response in renal cell carcinoma. JAMA Oncol 2019;5:1631–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Seliger B, Koehl U. Underlying mechanisms of evasion from NK cells as rationale for improvement of NK cell-based immunotherapies. Front Immunol 2022;13:910595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mazzaschi G, Facchinetti F, Missale G, Canetti D, Madeddu D, Zecca A, et al. The circulating pool of functionally competent NK and CD8+ cells predicts the outcome of anti-PD1 treatment in advanced NSCLC. Lung Cancer 2019;127:153–63. [DOI] [PubMed] [Google Scholar]
- 23. Jie HB, Schuler PJ, Lee SC, Srivastava RM, Argiris A, Ferrone S, et al. CTLA-4+ regulatory T cells increased in cetuximab-treated head and neck cancer patients suppress NK cell cytotoxicity and correlate with poor prognosis. Cancer Res 2015;75:2200–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kerdiles Y, Ugolini S, Vivier E. T cell regulation of natural killer cells. J Exp Med 2013;210:1065–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hu Z, Xu X, Wei H. The adverse impact of tumor microenvironment on NK-cell. Front Immunol 2021;12:633361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Morvan MG, Lanier LL. NK cells and cancer: you can teach innate cells new tricks. Nat Rev Cancer 2016;16:7–19. [DOI] [PubMed] [Google Scholar]
- 27. Beldi-Ferchiou A, Lambert M, Dogniaux S, Vély F, Vivier E, Olive D, et al. PD-1 mediates functional exhaustion of activated NK cells in patients with Kaposi sarcoma. Oncotarget 2016;7:72961–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Long EO, Kim HS, Liu D, Peterson ME, Rajagopalan S. Controlling natural killer cell responses: integration of signals for activation and inhibition. Annu Rev Immunol 2013;31:227–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bi J, Tian Z. NK cell exhaustion. Front Immunol 2017;8:760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hsu J, Hodgins JJ, Marathe M, Nicolai CJ, Bourgeois-Daigneault MC, Trevino TN, et al. Contribution of NK cells to immunotherapy mediated by PD-1/PD-L1 blockade. J Clin Invest 2018;128:4654–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Concha-Benavente F, Kansy B, Moskovitz J, Moy J, Chandran U, Ferris RL. PD-L1 mediates dysfunction in activated PD-1+ NK cells in head and neck cancer patients. Cancer Immunol Res 2018;6:1548–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sasidharan Nair V, Elkord E. Immune checkpoint inhibitors in cancer therapy: a focus on T-regulatory cells. Immunol Cell Biol 2018;96:21–33. [DOI] [PubMed] [Google Scholar]
- 33. Aksoylar HI, Boussiotis VA. PD-1. Nat Immunol 2020;21:1311–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Juliá EP, Mandó P, Rizzo MM, Cueto GR, Tsou F, Luca R, et al. Peripheral changes in immune cell populations and soluble mediators after anti-PD-1 therapy in non-small cell lung cancer and renal cell carcinoma patients. Cancer Immunol Immunother 2019;68:1585–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ottonello S, Genova C, Cossu I, Fontana V, Rijavec E, Rossi G, et al. Association between response to nivolumab treatment and peripheral blood lymphocyte subsets in patients with non-small cell lung cancer. Front Immunol 2020;11:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Khoja L, Kibiro M, Metser U, Gedye C, Hogg D, Butler MO, et al. Patterns of response to anti-PD-1 treatment: an exploratory comparison of four radiological response criteria and associations with overall survival in metastatic melanoma patients. Br J Cancer 2016;115:1186–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Niu C, Li M, Zhu S, Chen Y, Zhou L, Xu D, et al. PD-1-positive natural killer cells have a weaker antitumor function than that of PD-1-negative natural killer cells in lung cancer. Int J Med Sci 2020;17:1964–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Trefny MP, Kaiser M, Stanczak MA, Herzig P, Savic S, Wiese M, et al. PD-1+ natural killer cells in human non-small cell lung cancer can be activated by PD-1/PD-L1 blockade. Cancer Immunol Immunother 2020;69:1505–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Romero I, Garrido C, Algarra I, Chamorro V, Collado A, Garrido F, et al. MHC intratumoral heterogeneity may predict cancer progression and response to immunotherapy. Front Immunol 2018;9:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Aptsiauri N, Ruiz-Cabello F, Garrido F. The transition from HLA-I positive to HLA-I negative primary tumors: the road to escape from T-cell responses. Curr Opin Immunol 2018;51:123–32. [DOI] [PubMed] [Google Scholar]
- 41. Björkström NK, Riese P, Heuts F, Andersson S, Fauriat C, Ivarsson MA, et al. Expression patterns of NKG2A, KIR, and CD57 define a process of CD56dim NK-cell differentiation uncoupled from NK-cell education. Blood 2010;116:3853–64. [DOI] [PubMed] [Google Scholar]
- 42. Moretta L, Locatelli F, Pende D, Marcenaro E, Mingari MC, Moretta A. Killer Ig–like receptor-mediated control of natural killer cell alloreactivity in haploidentical hematopoietic stem cell transplantation. Blood 2011;117:764–71. [DOI] [PubMed] [Google Scholar]
- 43. Feils AS, Erbe AK, Birstler J, Kim K, Hoch U, Currie SL, et al. Associations between KIR/KIR-ligand genotypes and clinical outcome for patients with advanced solid tumors receiving BEMPEG plus nivolumab combination therapy in the PIVOT-02 trial. Cancer Immunol Immunother 2023;72:2099–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ishida Y, Nakashima C, Kojima H, Tanaka H, Fujimura T, Matsushita S, et al. Killer immunoglobulin-like receptor genotype did not correlate with response to anti-PD-1 antibody treatment in a Japanese cohort. Sci Rep 2018;8:15962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Thomas LM, Peterson ME, Long EO. Cutting edge: NK cell licensing modulates adhesion to target cells. J Immunol 2013;191:3981–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Palamarchuk AI, Alekseeva NA, Streltsova MA, Ustiuzhanina MO, Kobyzeva PA, Kust SA, et al. Increased susceptibility of the CD57− NK cells expressing kir2dl2/3 and NKG2C to iCasp9 gene retroviral transduction and the relationships with proliferative potential, activation degree, and death induction response. Int J Mol Sci 2021;22:13326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jonsson AH, Yokoyama WM. Natural killer cell tolerance licensing and other mechanisms. Adv Immunol 2009;101:27–79. [DOI] [PubMed] [Google Scholar]
- 48. Kim S, Sunwoo JB, Yang L, Choi T, Song YJ, French AR, et al. HLA alleles determine differences in human natural killer cell responsiveness and potency. Proc Natl Acad Sci U S A 2008;105:3053–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang W, Erbe AK, Alderson KA, Phillips E, Gallenberger M, Gan J, et al. Human NK cells maintain licensing status and are subject to killer immunoglobulin-like receptor (KIR) and KIR-ligand inhibition following ex vivo expansion. Cancer Immunol Immunother 2016;65:1047–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Romero JM, Aptsiauri N, Vazquez F, Cozar JM, Canton J, Cabrera T, et al. Analysis of the expression of HLA class I, proinflammatory cytokines and chemokines in primary tumors from patients with localized and metastatic renal cell carcinoma. Tissue Antigens 2006;68:303–10. [DOI] [PubMed] [Google Scholar]
- 51. Schwarzer A, Wolf B, Fisher JL, Schwaab T, Olek S, Baron U, et al. Regulatory T-cells and associated pathways in metastatic renal cell carcinoma (mRCC) patients undergoing DC-vaccination and cytokine-therapy. PLoS One 2012;7:e46600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Elkord E, Sharma S, Burt DJ, Hawkins RE. Expanded subpopulation of FoxP3+ T regulatory cells in renal cell carcinoma co-express Helios, indicating they could be derived from natural but not induced Tregs. Clin Immunol 2011;140:218–22. [DOI] [PubMed] [Google Scholar]
- 53. Santagata S, Napolitano M, D’Alterio C, Desicato S, Maro SD, Marinelli L, et al. Targeting CXCR4 reverts the suppressive activity of T-regulatory cells in renal cancer. Oncotarget 2017;8:77110–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kamada T, Togashi Y, Tay C, Ha D, Sasaki A, Nakamura Y, et al. PD-1+ regulatory T cells amplified by PD-1 blockade promote hyperprogression of cancer. Proc Natl Acad Sci U S A 2019;116:9999–10008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Woods DM, Ramakrishnan R, Laino AS, Berglund A, Walton K, Betts BC, et al. Decreased suppression and increased phosphorylated STAT3 in regulatory T cells are associated with benefit from adjuvant PD-1 blockade in resected metastatic melanoma. Clin Cancer Res 2018;24:6236–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gambichler T, Schröter U, Höxtermann S, Susok L, Stockfleth E, Becker JC. Decline of programmed death-1-positive circulating T regulatory cells predicts more favourable clinical outcome of patients with melanoma under immune checkpoint blockade. Br J Dermatol 2020;182:1214–20. [DOI] [PubMed] [Google Scholar]
- 57. Muto S, Owada Y, Inoue T, Watanabe Y, Yamaura T, Fukuhara M, et al. Clinical significance of expanded Foxp3+ Helios− regulatory T cells in patients with non-small cell lung cancer. Int J Oncol 2015;47:2082–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lam AJ, Uday P, Gillies JK, Levings MK. Helios is a marker, not a driver, of human Treg stability. Eur J Immunol 2022;52:75–84. [DOI] [PubMed] [Google Scholar]
- 59. Zhang Y, Zhang J, Shi Y, Shen M, Lv H, Chen S, et al. Differences in maturation status and immune phenotypes of circulating Helios+ and Helios− Tregs and their disrupted correlations with monocyte subsets in autoantibody-positive T1D individuals. Front Immunol 2021;12:628504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kim YC, Bhairavabhotla R, Yoon J, Golding A, Thornton AM, Tran DQ, et al. Oligodeoxynucleotides stabilize Helios-expressing Foxp3+ human T regulatory cells during in vitro expansion. Blood 2012;119:2810–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Himmel ME, MacDonald KG, Garcia RV, Steiner TS, Levings MK. Helios+ and Helios− cells coexist within the natural FOXP3+ T regulatory cell subset in humans. J Immunol 2013;190:2001–8. [DOI] [PubMed] [Google Scholar]
- 62. Raffin C, Pignon P, Celse C, Debien E, Valmori D, Ayyoub M. Human memory Helios− FOXP3+ regulatory T cells (Tregs) encompass induced Tregs that express Aiolos and respond to IL-1β by downregulating their suppressor functions. J Immunol 2013;191:4619–27. [DOI] [PubMed] [Google Scholar]
- 63. Bin Dhuban K, d’Hennezel E, Nashi E, Bar-Or A, Rieder S, Shevach EM, et al. Coexpression of TIGIT and FCRL3 identifies Helios+ human memory regulatory T cells. J Immunol 2015;194:3687–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Giraldo NA, Becht E, Vano Y, Petitprez F, Lacroix L, Validire P, et al. Tumor-infiltrating and peripheral blood T-cell immunophenotypes predict early relapse in localized clear cell renal cell carcinoma. Clin Cancer Res 2017;23:4416–28. [DOI] [PubMed] [Google Scholar]
- 65. Whiteside TL. The role of regulatory T cells in cancer immunology. Immunotargets Ther 2015;4:159–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lin F, Hu X, Zhang Y, Ye S, Gu Y, Yan B, et al. Upregulated TIGIT+ and Helios+ regulatory T cell levels in bronchoalveolar lavage fluid of NSCLC patients. Mol Immunol 2022;147:40–9. [DOI] [PubMed] [Google Scholar]
- 67. Timperi E, Pacella I, Schinzari V, Focaccetti C, Sacco L, Farelli F, et al. Regulatory T cells with multiple suppressive and potentially pro-tumor activities accumulate in human colorectal cancer. Oncoimmunology 2016;5:e1175800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Thornton AM, Korty PE, Tran DQ, Wohlfert EA, Murray PE, Belkaid Y, et al. Expression of Helios, an ikaros transcription factor family member, differentiates thymic-derived from peripherally induced Foxp3+ T regulatory cells. J Immunol 2010;184:3433–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Di Maro S, Di Leva FS, Trotta AM, Brancaccio D, Portella L, Aurilio M, et al. Structure–activity relationships and biological characterization of a novel, potent, and serum stable C-X-C chemokine receptor type 4 (CXCR4) antagonist. J Med Chem 2017;60:9641–52. [DOI] [PubMed] [Google Scholar]
- 70. Santagata S, Rea G, Bello AM, Capiluongo A, Napolitano M, Desicato S, et al. Targeting CXCR4 impaired T regulatory function through PTEN in renal cancer patients. Br J Cancer 2024;130:2016–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1. Antibodies used for flow cytometry Supplementary Table S2. mRCC-REV patients characteristics Supplementary Table S3. HDs characteristics Supplementary Table S4. Representativeness of Study Participants Supplementary Table S5. Univariate analysis of peripheral immune cell subsets between HD and mRCC-REV groups Supplementary Table S6. Longitudinal evaluation of peripheral immune cell subsets in mRCC-REV patients Supplementary Table S7. Differential distribution of peripheral immune cell subsets between R and NR groups (T0) in mRCC-REV patients Supplementary Table S8. Univariate analysis of peripheral immune cell subsets between R and NR groups including early death patients (T0) in mRCC-REV patients Supplementary Table S9. Cut-off value prediction with ROC curve for continuous variable at basal level and response to nivolumab after 3 months as classification variable Supplementary Table S10. Association between the KIR2DL2/DL3+NKs/Helios+Tregs values and response to nivolumab treatment Supplementary Table S11. Differential distribution of peripheral immune cell subsets between R and NR groups (T1) in mRCC-REV patients Supplementary Table S12. Cut-off value prediction with ROC curve for continuous variable at T1 and response to nivolumab after 3 months as classification variable
Supplementary Figure S1. Gating strategy for identification of Tregs and NKs in peripheral blood.
Supplementary Figure S2. Correlation between age and pretreatment NKs and Tregs immune subsets cells in mRCC-REV patients and HDs.
Supplementary Figure S3. Pretreatment NKs and Tregs in mRCC-REV patients
Supplementary Figure S4. Pretreatment NKs co-expression of PD-1 and KIR2DL1or KIR2DL2/DL3 in mRCC-REV patients.
Supplementary Figure S5. Nivolumab did not significantly affect peripheral NKs and Tregs
Supplementary Figure S6. Swimmer’s plot of nivolumab-treated mRCC patients.
Supplementary Figure S7. Pretreatment NKs and Tregs subpopulations in mRCC-REV Long Responder (LR) patients vs Not-Responders (NR).
Supplementary Figure S8. .632+ cross validation bootstrap ROC curves and the optimum pretreatment cut-off values of KIR2DL2/DL3+NKs and Helios+Tregs to determine the outcome of REV patients
Supplementary Figure S9. Association between the KIR2DL2/DL3+NKs/Helios+Tregs values and response to nivolumab treatment.
Supplementary Figure S10. .632+ cross validation bootstrap ROC curves and the optimum T1 cut-off values of CD3+ and KIR2DL2/DL3+NKs to determine the outcome of REV patients
Supplementary Figure S11. HLA class I in primary RCC from REV patients
Supplementary Figure S12. Correlation between pretreatment of PD-1+Tregs and Helios+Tregs in Responder and Not-Responder patients.
Supplementary Figure S13. T0-T1 values (after one month of nivolumab) between PD-1+ Tregs and Helios+Tregs in Responder and Not Responder patients
Data Availability Statement
The data are openly available at https://zenodo.org/records/11394968 and from the corresponding author upon reasonable request.