Abstract
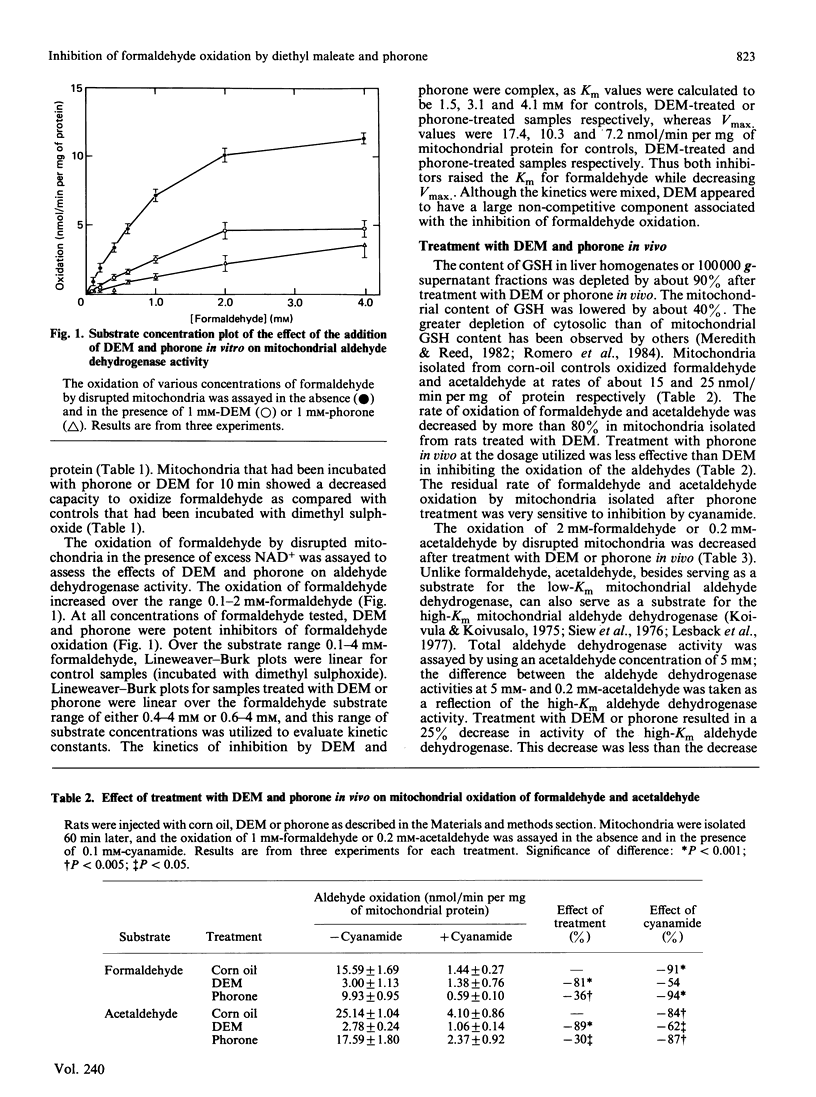
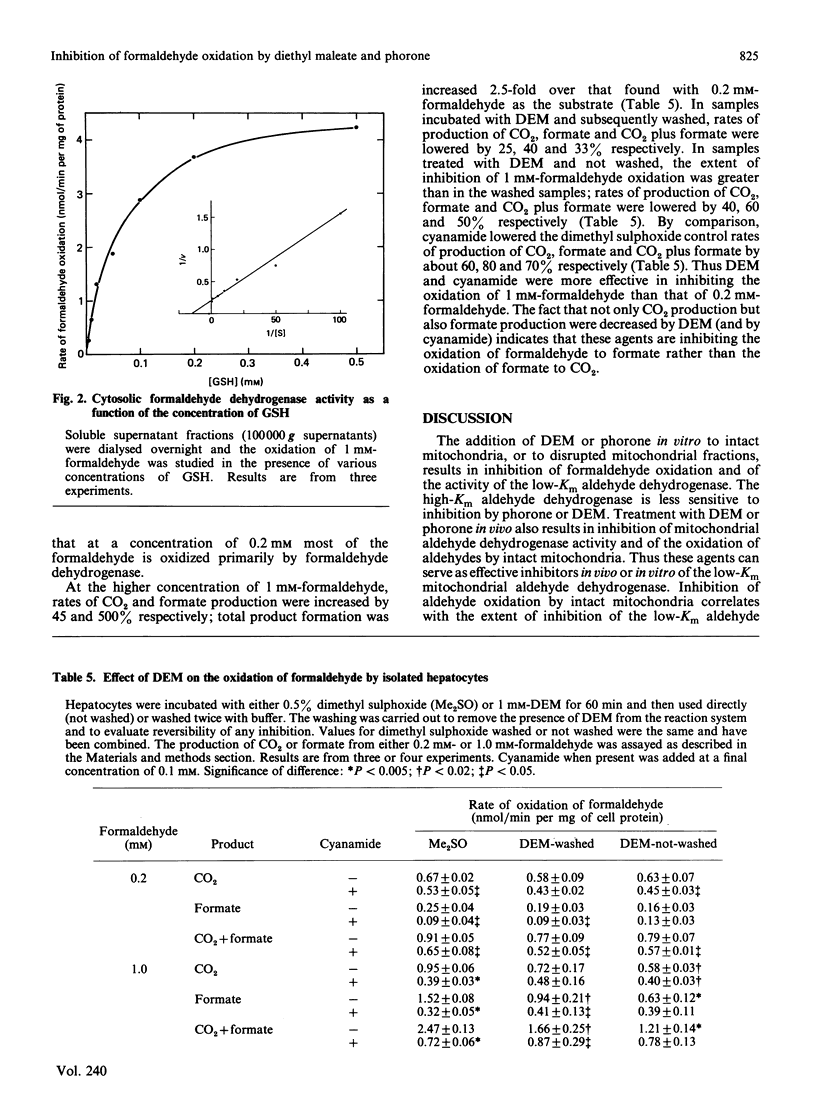
Formaldehyde can be oxidized primarily by two different enzymes, the low-Km mitochondrial aldehyde dehydrogenase and the cytosolic GSH-dependent formaldehyde dehydrogenase. Experiments were carried out to evaluate the effects of diethyl maleate or phorone, agents that deplete GSH from the liver, on the oxidation of formaldehyde. The addition of diethyl maleate or phorone to intact mitochondria or to disrupted mitochondrial fractions produced inhibition of formaldehyde oxidation. The kinetics of inhibition of the low-Km mitochondrial aldehyde dehydrogenase were mixed. Mitochondria isolated from rats treated in vivo with diethyl maleate or phorone had a decreased capacity to oxidize either formaldehyde or acetaldehyde. The activity of the low-Km, but not the high-Km, mitochondrial aldehyde dehydrogenase was also inhibited. The production of CO2 plus formate from 0.2 mM-[14C]formaldehyde by isolated hepatocytes was only slightly inhibited (15-30%) by incubation with diethyl maleate or addition of cyanamide, suggesting oxidation primarily via formaldehyde dehydrogenase. However, the production of CO2 plus formate was increased 2.5-fold when the concentration of [14C]formaldehyde was raised to 1 mM. This increase in product formation at higher formaldehyde concentrations was much more sensitive to inhibition by diethyl maleate or cyanamide, suggesting an important contribution by mitochondrial aldehyde dehydrogenase. Thus diethyl maleate and phorone, besides depleting GSH, can also serve as effective inhibitors in vivo or in vitro of the low-Km mitochondrial aldehyde dehydrogenase. Inhibition of formaldehyde oxidation by these agents could be due to impairment of both enzyme systems known to be capable of oxidizing formaldehyde. It would appear that a critical amount of GSH, e.g. 90%, must be depleted before the activity of formaldehyde dehydrogenase becomes impaired.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bhatt H. S., Combes B. The effect of glutathione depletion on 14CO2 evolution from [14C]methyl-labeled aminopyrine in intact rats. Hepatology. 1985 Jul-Aug;5(4):615–621. doi: 10.1002/hep.1840050416. [DOI] [PubMed] [Google Scholar]
- Billings R. E., Tephly T. R. Studies on methanol toxicity and formate metabolism in isolated hepatocytes. The role of methionine in folate-dependent reactions. Biochem Pharmacol. 1979 Oct 1;28(19):2985–2991. doi: 10.1016/0006-2952(79)90598-7. [DOI] [PubMed] [Google Scholar]
- Boyland E., Chasseaud L. F. The effect of some carbonyl compounds on rat liver glutathione levels. Biochem Pharmacol. 1970 Apr;19(4):1526–1528. doi: 10.1016/0006-2952(70)90075-4. [DOI] [PubMed] [Google Scholar]
- Cederbaum A. I., Dicker E., Rubin E. Transfer and reoxidation of reducing equivalents as the rate-limiting steps in the oxidation of ethanol by liver cells isolated from fed and fasted rats. Arch Biochem Biophys. 1977 Oct;183(2):638–646. doi: 10.1016/0003-9861(77)90398-8. [DOI] [PubMed] [Google Scholar]
- Cederbaum A. I. The effect of cyanamide on acetaldehyde oxidation by isolated rat liver mitochondria and on the inhibition of pyruvate oxidation by acetaldehyde. Alcohol Clin Exp Res. 1981 Jan;5(1):38–44. doi: 10.1111/j.1530-0277.1981.tb04862.x. [DOI] [PubMed] [Google Scholar]
- Cinti D. L., Keyes S. R., Lemelin M. A., Denk H., Schenkman J. B. Biochemical properties of rat liver mitochondrial aldehyde dehydrogenase with respect to oxidation of formaldehyde. J Biol Chem. 1976 Mar 25;251(6):1571–1577. [PubMed] [Google Scholar]
- DEITRICH R. A., HELLERMAN L. Diphosphopridine nucleotide-linked aldehyde dehydrogenase. II. Inhibitors. J Biol Chem. 1963 May;238:1683–1689. [PubMed] [Google Scholar]
- Dicker E., Cederbaum A. I. Effect of acetaldehyde and cyanamide on the metabolism of formaldehyde by hepatocytes, mitochondria, and soluble supernatant from rat liver. Arch Biochem Biophys. 1984 Jul;232(1):179–188. doi: 10.1016/0003-9861(84)90533-2. [DOI] [PubMed] [Google Scholar]
- Dicker E., Cederbaum A. I. Inhibition of mitochondrial aldehyde dehydrogenase and acetaldehyde oxidation by the glutathione-depleting agents diethylmaleate and phorone. Biochim Biophys Acta. 1985 Nov 22;843(1-2):107–113. doi: 10.1016/0304-4165(85)90056-x. [DOI] [PubMed] [Google Scholar]
- Dicker E., Cederbaum A. I. Inhibition of the oxidation of acetaldehyde and formaldehyde by hepatocytes and mitochondria by crotonaldehyde. Arch Biochem Biophys. 1984 Oct;234(1):187–196. doi: 10.1016/0003-9861(84)90340-0. [DOI] [PubMed] [Google Scholar]
- Eckfeldt J., Mope L., Takio K., Yonetani T. Horse liver aldehyde dehydrogenase. Purification and characterization of two isozymes. J Biol Chem. 1976 Jan 10;251(1):236–240. [PubMed] [Google Scholar]
- Goodman J. I., Tephly T. R. A comparison of rat and human liver formaldehyde dehydrogenase. Biochim Biophys Acta. 1971 Dec 21;252(3):489–505. doi: 10.1016/0304-4165(71)90152-8. [DOI] [PubMed] [Google Scholar]
- Jones D. P., Thor H., Andersson B., Orrenius S. Detoxification reactions in isolated hepatocytes. Role of glutathione peroxidase, catalase, and formaldehyde dehydrogenase in reactions relating to N-demethylation by the cytochrome P-450 system. J Biol Chem. 1978 Sep 10;253(17):6031–6037. [PubMed] [Google Scholar]
- Kitson T. M. The disulfiram--ethanol reaction: a review. J Stud Alcohol. 1977 Jan;38(1):96–113. doi: 10.15288/jsa.1977.38.96. [DOI] [PubMed] [Google Scholar]
- Koivula T., Koivusalo M. Different forms of rat liver aldehyde dehydrogenase and their subcellular distribution. Biochim Biophys Acta. 1975 Jul 27;397(1):9–23. doi: 10.1016/0005-2744(75)90174-6. [DOI] [PubMed] [Google Scholar]
- Krebs H. A., Hems R., Tyler B. The regulation of folate and methionine metabolism. Biochem J. 1976 Aug 15;158(2):341–353. doi: 10.1042/bj1580341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku R. H., Billings R. E. Relationships between formaldehyde metabolism and toxicity and glutathione concentrations in isolated rat hepatocytes. Chem Biol Interact. 1984 Sep 1;51(1):25–36. doi: 10.1016/0009-2797(84)90017-6. [DOI] [PubMed] [Google Scholar]
- Lebsack M. E., Petersen D. R., Collins A. C., Anderson A. D. Preferential inhibition of the low Km aldehyde dehydrogenase activity by pargyline. Biochem Pharmacol. 1977 Jun 15;26(12):1151–1154. doi: 10.1016/0006-2952(77)90060-0. [DOI] [PubMed] [Google Scholar]
- Meredith M. J., Reed D. J. Status of the mitochondrial pool of glutathione in the isolated hepatocyte. J Biol Chem. 1982 Apr 10;257(7):3747–3753. [PubMed] [Google Scholar]
- NASH T. The colorimetric estimation of formaldehyde by means of the Hantzsch reaction. Biochem J. 1953 Oct;55(3):416–421. doi: 10.1042/bj0550416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poli G., Dianzani M. U., Cheeseman K. H., Slater T. F., Lang J., Esterbauer H. Separation and characterization of the aldehydic products of lipid peroxidation stimulated by carbon tetrachloride or ADP-iron in isolated rat hepatocytes and rat liver microsomal suspensions. Biochem J. 1985 Apr 15;227(2):629–638. doi: 10.1042/bj2270629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson R. J., Murphy S. D. Effect of glutathione depletion on tissue deposition of methylmercury in rats. Toxicol Appl Pharmacol. 1975 Mar;31(3):505–519. doi: 10.1016/0041-008x(75)90274-4. [DOI] [PubMed] [Google Scholar]
- Romero F. J., Soboll S., Sies H. Mitochondrial and cytosolic glutathione after depletion by phorone in isolated hepatocytes. Experientia. 1984 Apr 15;40(4):365–367. doi: 10.1007/BF01952555. [DOI] [PubMed] [Google Scholar]
- STRITTMATTER P., BALL E. G. Formaldehyde dehydrogenase, a glutathionedependent enzyme system. J Biol Chem. 1955 Mar;213(1):445–461. [PubMed] [Google Scholar]
- Savenije-Chapel E. M., Noordhoek J. Metabolism of formaldehyde during in vitro drug demethylation. Biochem Pharmacol. 1980 Jul 15;29(14):2023–2029. doi: 10.1016/0006-2952(80)90487-6. [DOI] [PubMed] [Google Scholar]
- Siew C., Deitrich R. A., Erwin V. G. Localization and characteristics of rat liver mitochondrial aldehyde dehydrogenases. Arch Biochem Biophys. 1976 Oct;176(2):638–649. doi: 10.1016/0003-9861(76)90208-3. [DOI] [PubMed] [Google Scholar]
- Uotila L., Koivusalo M. Formaldehyde dehydrogenase from human liver. Purification, properties, and evidence for the formation of glutathione thiol esters by the enzyme. J Biol Chem. 1974 Dec 10;249(23):7653–7663. [PubMed] [Google Scholar]
- Uotila L., Mannervik B. A steady-state-kinetic model for formaldehyde dehydrogenase from human liver. A mechanism involving NAD+ and the hemimercaptal adduct of glutathione and formaldehyde as substrates and free glutathione as an allosteric activator of the enzyme. Biochem J. 1979 Mar 1;177(3):869–878. doi: 10.1042/bj1770869. [DOI] [PMC free article] [PubMed] [Google Scholar]


