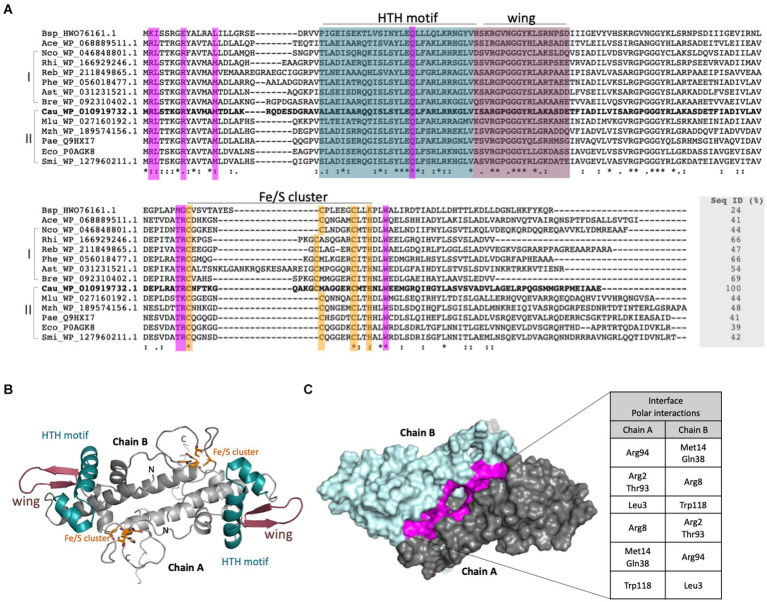
Figure 2.
Sequential and structural characterization of the C. crescentus IscR. (A). Structural alignment of putative IscR proteins from selected bacteria was made with Expresso from webserver T-Coffee (Notredame et al., 2000). Bsp_HWO_76161.1: Bacillus sp.; Ace_WP_068889511.1: Acinetobacter celticus; Nco_WP_046848801.1: Nitrosomonas communis; Rhi_WP_166929246.1: Rhizomicrobium electricum; Reb_WP_211849865.1: Roseomonas eburnea; Phe_WP_056018477.1: Phenylobacterium sp.; Ast_WP_031231521.1: Asticcacaulis sp. YBE204; Bre_WP_092310402.1 Brevundimonas viscosa; Cau_YP_002517315 Caulobacter crescentus NA1000; Mlu_WP_027160192.1 Methylobacter luteus; Mzh_WP_189574156.1 Marinobacter zhanjiangensis; Pae_NC_002516.2: Pseudomonas aeruginosa; Eco_NP_417026.1 Escherichia coli; Smi_WP_127960211.1: Serratia microhaemolytica. I and II: Sequences were classified in two groups according to the phylogenetic tree. Residues belonging to the HTH motif and wing are colored in deep cyan and dark purple, respectively. Residues involved in the Fe-S cluster binding (3 Cys and 1 His) are shown in orange and residues that form an interface between both monomers are shown in purple. (B) Model of the IscR generated with AlphaFold2. The dimer is shown in cartoon representation with the motifs and residues colored according to (A). (C) Dimer of IscR in surface mode highlighting the interface between the monomers in purple. The polar interactions performed are described in the table.