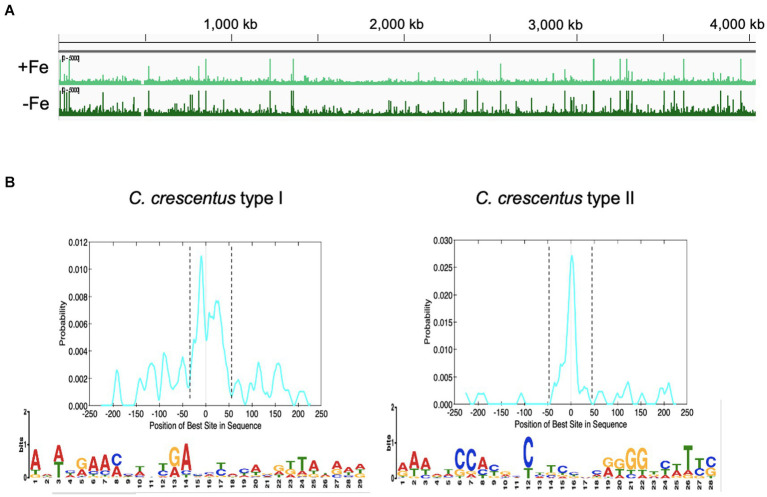
Figure 8.
Mapping the IscR-binding sites in the C. crescentus genome. (A) Peaks of DNA reads distributed along the C. crescentus genome obtained in the ChIP-seq experiment from cultures in PYE medium (+Fe) or PYE + DP (−Fe). (B) Motifs enriched in the regions containing the IscR-binding sites. FASTA files of 500-pb sequences containing the peaks summits centralized were used as input in the MEME-ChIP (Motif Analysis of Large Nucleotide Datasets) program of the MEME suite (58). The localization of the motif in the input sequences was found by CentriMo in the MEME suite, within the dashed lines (graphs at the top). Best score (1.8e-15) Type I motif identified in 43/184 DNA regions that were not differentially bound by IscR in the presence of DP. Best score (6.3e-016) Type II motif identified in 36/52 DNA regions that were preferentially bound by IscR in the presence of DP. IscR binding motifs were generated using WebLogo.