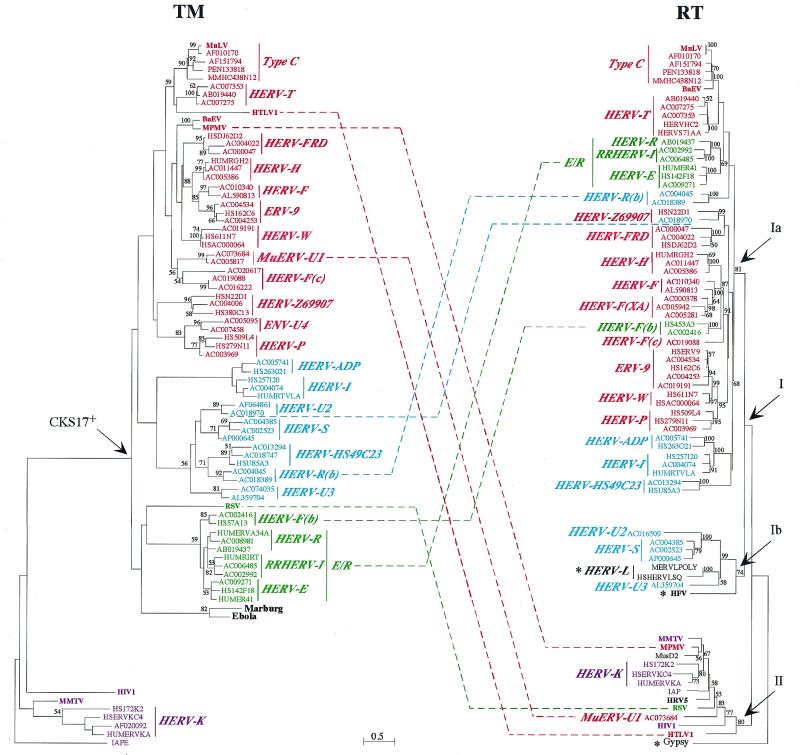
FIG. 5.
TM and RT protein phylogenetic trees. Both trees were determined by the neighbor-joining method with horizontal branch lengths proportional to the degree of divergence between the sequences (common scale for both trees). Vertical bars are only for presentation, with a few of them lengthened to highlight the major groups of sequences. The TM tree (left) is presented with the IAPE sequence (a murine retrotransposon envelope) as an outgroup, and the RT tree (right) is presented with Gypsy (a Drosophila melanogaster retrotransposon) as an outgroup. Percent bootstrap values obtained from 100 replicates are indicated on the branches only when they are >50%. Asterisks in the RT tree are for families devoid of the TM region (HERV-L family) or for which the TM could not be aligned (HFV and Gypsy). Major chimerisms are indicated by dotted lines between sequences from both trees. All identifiers and ERV families names are the same as in the TM alignment (Fig. 3). The few RT-only sequences are as follow: MERVLPOLY and HSHERVLSQ, the murine and human prototypic ERV-L sequences, respectively; HFV (HSPGAGPOL); MusD2 (AF246633) (27), a new mouse ERV devoid of the env region; HRV5 (HRU46939); IAP (AC006584); and Gypsy (AF033821). Within the HERV-T family, HERVHC2 and HUMS71AA (in red) are two known HERV sequences, but they carry internal deletions in the PBS (among others) which had precluded the definite naming of the family. For the sequences exhibiting large deletions in the TM (Fig. 3) and thus not included in the TM tree, a phylogeny calculated on reduced TM alignments led to the expected conclusions that the HERV-F(XA) sequences (AC000378 and AC005942) branch with the HERV-F family, the AC016509 sequence branches with the HERV-U2 family, and the AC007204 sequence branches with the HERV-U3 family.