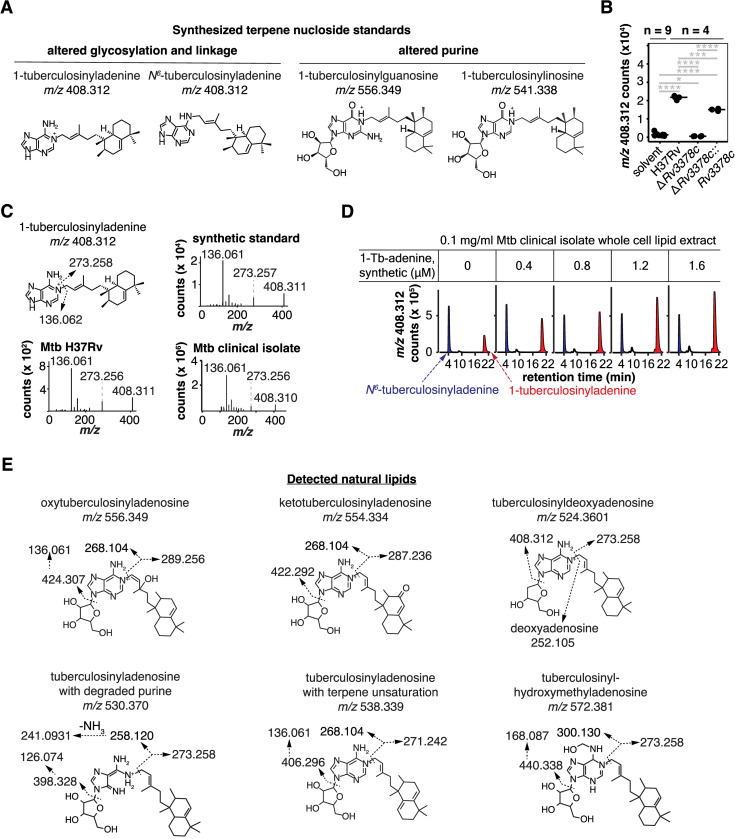
Fig 3. Unknown Rv3377-3378c-dependent lipids were identified as new terpene nucleoside family members.
(A) Structures of synthetic molecules used to analyze natural compounds. (B) Intensities of ion chromatograms corresponding to m/z 408.312, the most abundant non-TbAd lipid found in the differential abundance analysis. Significant pairwise t tests after Benjamini–Hochberg adjustment are indicated (*: p < 0.05, **: p < 0.01, ***: p < 0.001, ****: p < 0.0001). Raw data for these measurements were provided in S1 Data. (C) CID-MS of m/z 408.312 showed fragmentation patterns diagnostic of 1-tuberculosinyladenine. The chemical structure with fragmentation shows calculated masses while spectra show observed masses. (D) Mtb clinical isolate M0014870-1 total lipid extracts were spiked with synthetic 1-tuberculosinyladenine, which showed co-elution with natural 1-tuberculosinyladenine and established its chemical identity and absolute yield in Mtb. (E) Annotated fragments from CID-MS established structures of 6 previously unknown terpene nucleosides where the calculated masses are shown. Collision localized modifications to the ribose, adenosine, or terpene but linkage within the moiety was inferred based on known analogous compounds.