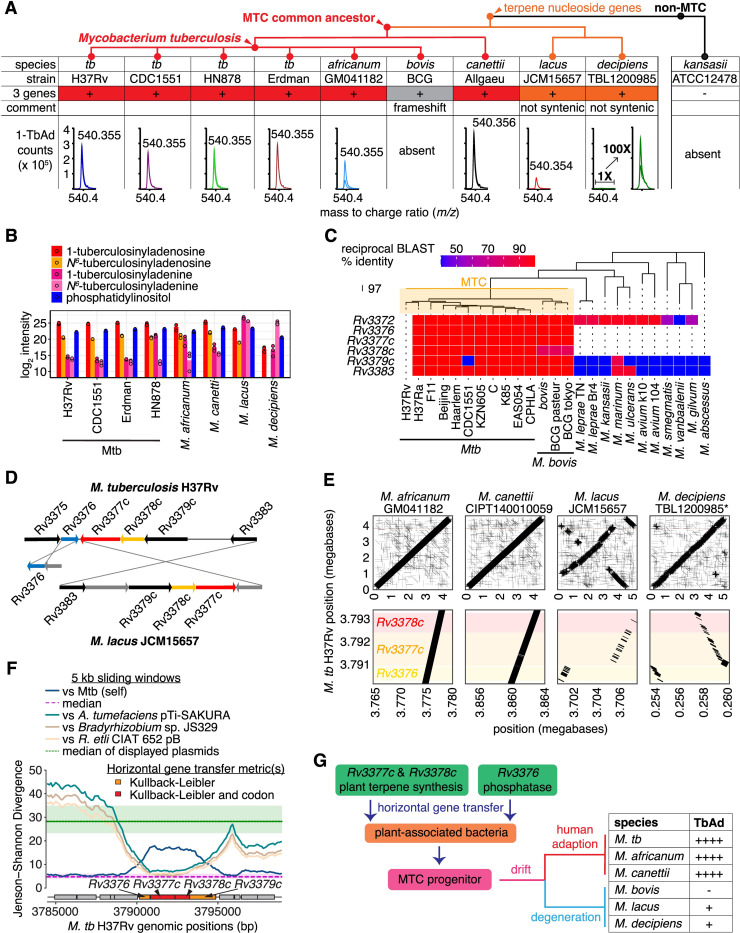
Fig 5. The timing and origin of horizontal transfer of biosynthetic genes for terpene nucleosides.
(A) Mass spectrometry tested 1-TbAd production and abundance in MTC species that contained the three-gene locus. Positive mode extracted ion chromatograms for 1-TbAd show lipid counts near m/z 540.354 (observed mass shown) at a retention time of approximately 23 min in samples at 1 mg/ml total lipid. The lack of 1-TbAd in M. bovis and M. kansasii was documented previously [5,12]. A cladogram of arbitrary branch lengths reflected plausible organization [33]. (B) Peak intensity of a predominant membrane lipid, phosphatidylinositol, along with 1- and N6-TbAd, and 1- and N6-tuberculosinyladenine were measured among a panel of 8 MTC strains and species. (C) Reciprocal BLAST hit scores versus H37Rv are shown as a heatmap for the terpene nucleoside biosynthetic genes Rv3376, Rv3377c, and Rv3378c along with flanking genes. The neighbor-joining species dendrogram was based on whole-genome presence/absence of orthogroups. (D) Locus organization in M. lacus is rearranged relative to Mtb H37Rv. (E) Synteny of Mycobacterium species is shown for the whole genome and Rv3376-8c locus as dot plots compared to the reference Mtb H37Rv genome. The M. decipiens TBL1200985 genome is not fully assembled; hence, genome positions were inferred by scaffolding using the Mtb H37Rv genome. (F) Jenson–Shannon divergence profiles of Mtb H37Rv comparing to Mtb H37Rv genome itself or to DNA sequences from other bacteria using a 5 kb sliding window with 100 bp step. The gene schematic is colored according to Kullback–Leibler divergence. (G) Schematic of gene acquisition shows divergence and function. The data for Fig 5B and 5C can be found in S1 Data.