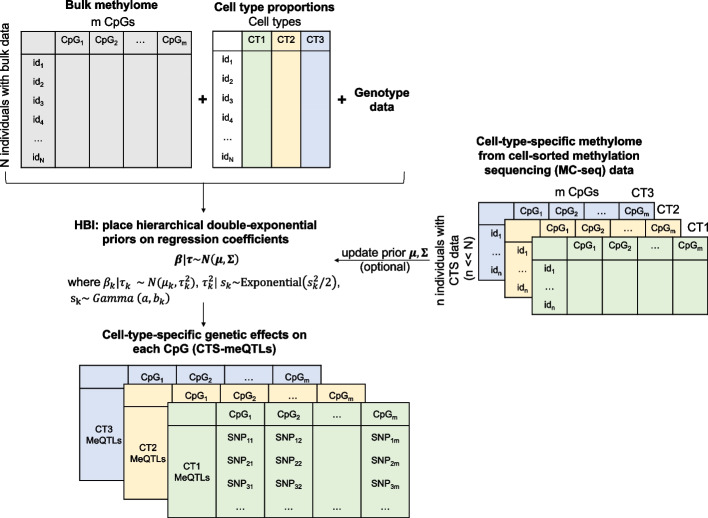
Fig. 1.
Overview of the hierarchical Bayesian interaction model (HBI) to infer cell type-specific (CTS) meQTLs. With bulk methylation data and cell type proportions (we present an example of three cell types: CT1, CT2, CT3), HBI employs an interaction model with sparse hierarchical priors placed on the regression coefficients for the interaction terms. If the CTS DNA methylation data (in our case, generated by methylation capture-sequencing using cells sorted from PBMC using flow cytometry) are available for a small group of samples, HBI will further incorporate the information into priors to refine the estimates for CTS genetic effects in the larger-scale bulk samples