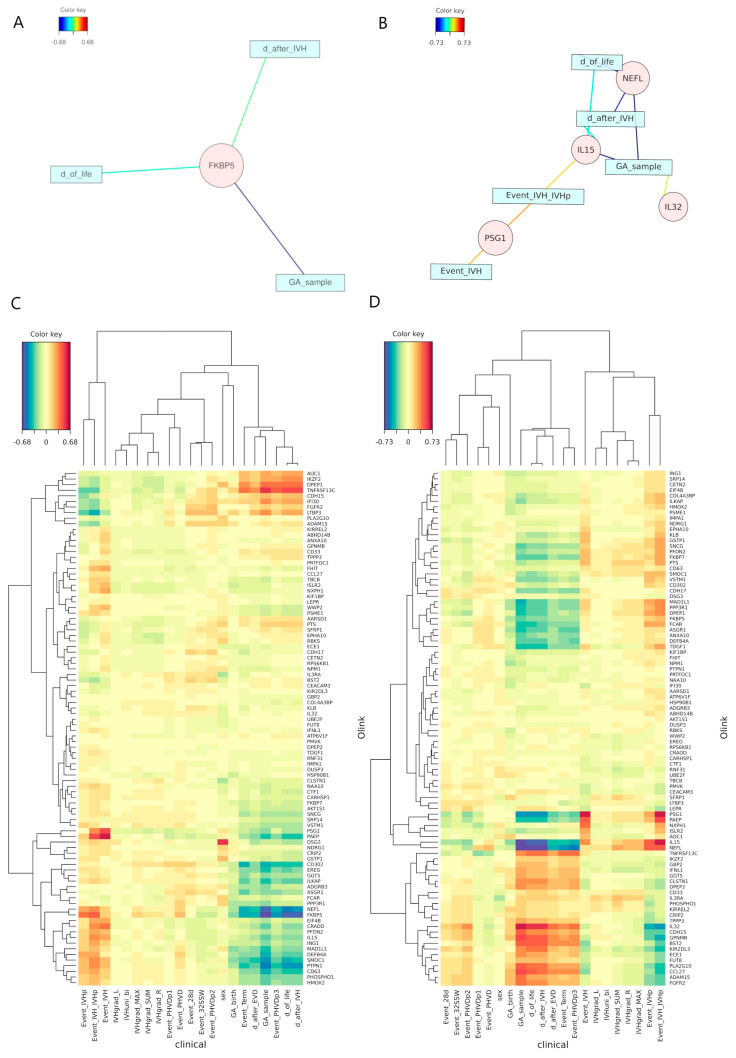
Figure 2.
(A)-visualization of the relevance associations network based on the rCCA between distinct clinical datasets characterizing the samples and the Olink measurements based on the urine data. The applied correlation threshold for inclusion in both networks was set to 0.6. (B)-visualization of the relevance associations network based on the rCCA between distinct clinical data sets characterizing the samples and the Olink measurements based on the serum data. (C)-heatmap of the rCCA between distinct clinical datasets characterizing the samples and the Olink measurements based on the urine data. The dendrogram in the heatmap was computed with the complete linkage method to find similar clusters based on the Euclidean distance. (D)-heatmap of the rCCA between distinct clinical datasets characterizing the samples and the Olink measurements based on the serum data. The dendrogram in the heatmap was computed with the complete linkage method to find similar clusters based on the Euclidean distance. All clinical data beginning with “Event_” indicate the samples belonging to a specific timeframe; these timeframes are highly correlated to features such as “day of life”, “day after IVH” or “GA of the sample”.