Figure 1.
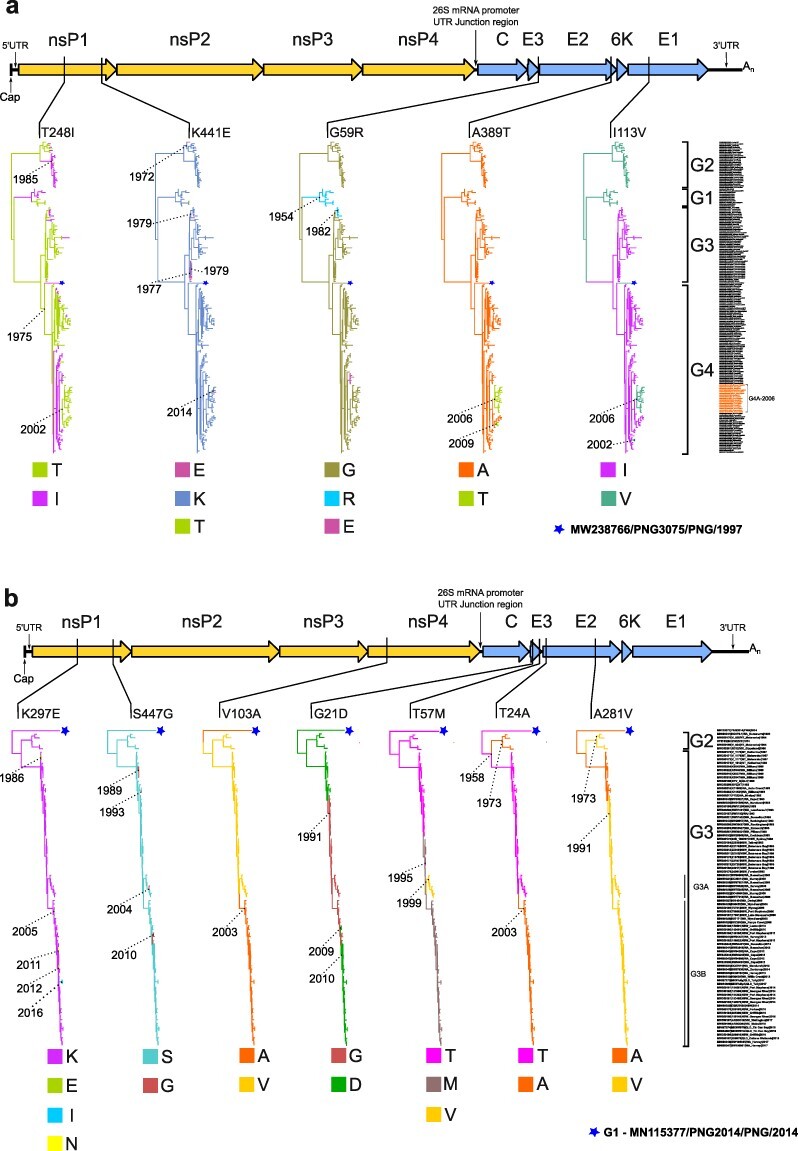
Alphavirus genome map and discrete-trait representation indicating genomic locations and lineage placement of Ross River virus and Barmah Forest virus convergent mutations. The schematic of the alphavirus genome at the top of each Fig. [A, Ross River virus (RRV) and B, Barmah Forest virus (BFV)] shows relative positions of convergent aa mutations within coding nonstructural (nsP1 to nsP4) or structural genes (C, E3, E2, 6 K, and E1). Each mutation is then further mapped below in individual maximum clade credibility (MCC) trees using their Markov rewards where branch color denotes the inferred ancestral state according to the amino acid named under each tree. Years are provided to denote the age of key nodes in the MCC trees. The taxa names and genotype groupings (RRV:G1 to G4, and BFV:G1 to G3) used to construct the MCC trees are also provided on the right side of each A and B sub-figure relative to their corresponding tree tip position. In Fig. 1a, the isolates belonging to the RRV G4A-2006 clade are highlighted on the figure (also summarized in Supplementary Table S8). The monophyletic PNG clade consisting of a single RRV PNG 1997 isolate, PNG3075 (MW238766) (Michie et al. 2021) in Fig. 1a, or BFV PNG 2014 isolate, PNG2014 (MN115377) (Caly et al. 2019) in Fig. 1b, are represented by stars.
