Figure 3.
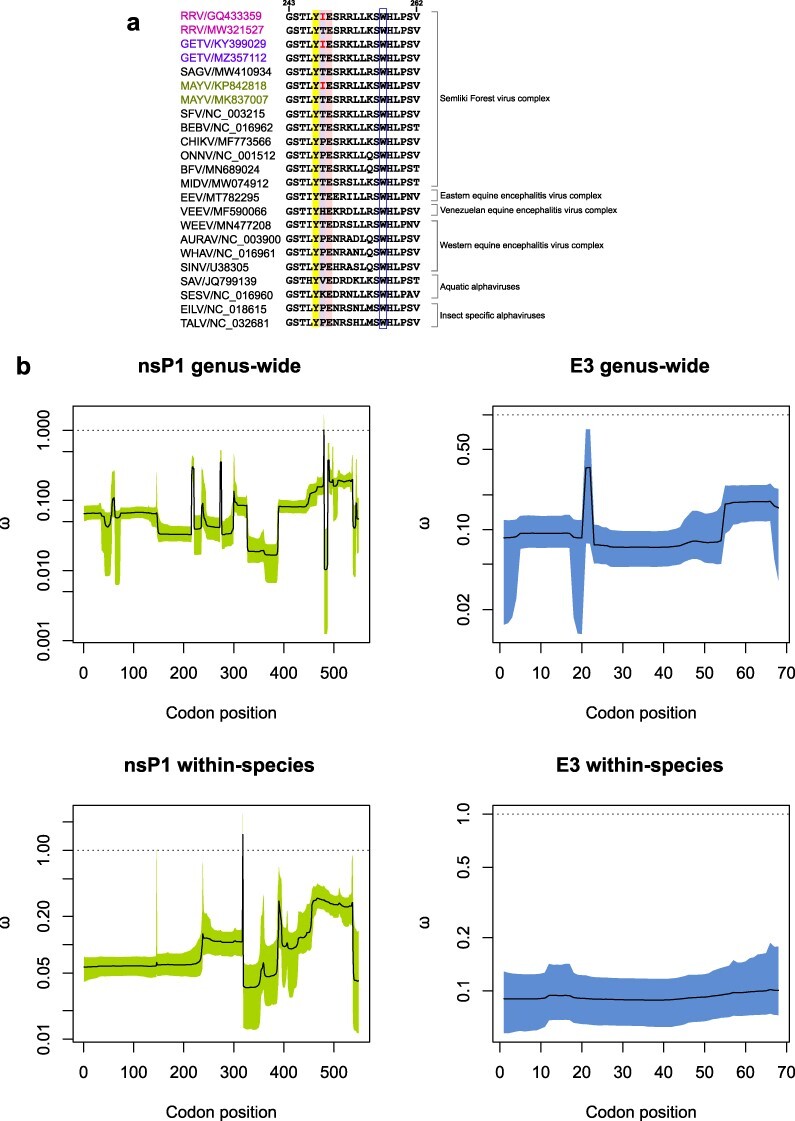
Protein sequence alignment of alphavirus nsP1 β11/αg region and GenomegaMap estimates of the nsP1 and E3 genus-wide and within-species dN/dS ratios ω. (A) Protein sequence alignment comparing RRV nsP1 residues G243 to V262 [previously reported as an amphipathic α-helix membrane binding peptide and encompassing the recently defined β11/αg region (Jones et al. 2021)] with cognate sequences from other representative Alphavirus species. The key residues, Y247 and E249, which are conserved in alphaviruses and have been identified as important for nsP1 methyltransferase activity, are highlighted in colour together with the RRV mutated residue at site 248. The conserved W257 site also considered an important membrane anchor for nsP1, is indicated in the open blue box. (B) GenomegaMap output plots. Solid lines (posterior medians) and shaded regions (95% credibility intervals) for the GenomegaMap point estimates for nsP1 and E3 genus-wide and within-species dN/dS ratios ω are shown. The dotted line (ω of 1) indicates the threshold for neutral selection.
