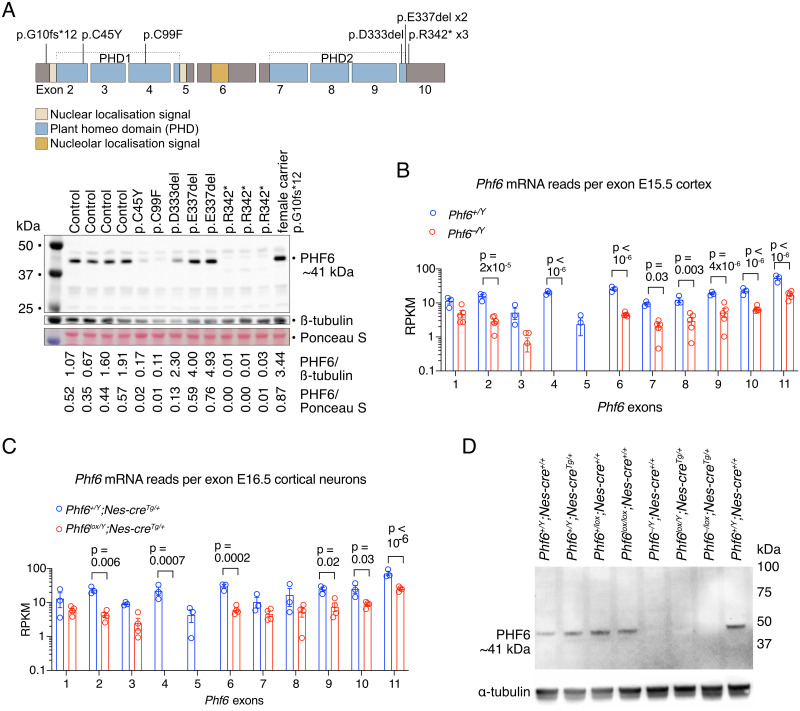
Fig 1. PHF6 protein levels in cells from BFLS individuals with PHF6 mutations, mRNA and protein levels in the cortex and cortical neurons of Phf6 deleted mice.
(A) PHF6 protein expression is reduced in cell lines derived from lymphocytes of individuals affected by BFLS. Schematic drawing of the protein domains and coding exon structure of the PHF6 gene with BFLS variants indicated and Western blot showing expression of PHF6 migrating at approximately 41 kDa in 8 males affected by BFLS, one female carrier and four unaffected males (control). Protein expression in individuals with the PHF6 p.C45Y, p.C99F and p.R342* variants was reduced, while the p.D333del and p.E337del variants are expressed at a similar or even higher level to the four unaffected individuals (control). Note the band migrating at approximately 38.5 kDa in the patients with the p.R342* variant indicating that the truncated protein is likely to be expressed at a low abundance. Details on individuals included in this figure are in S1 Table. (B, C) RNA-sequencing reads as reads per kilobase per one million reads in the sequencing library (RPKM) mapping to Phf6 exons in Phf6+/Y vs. Phf6–/Y mouse E15.5 developing cortex (B) and Phf6+/Y;Nes-creTg/+ vs. Phf6lox/Y;Nes-creTg/+ mouse E16.5 cortical neurons (C). (D) Detection of PHF6 protein by western immunoblotting in E15.5 mouse brains of Phf6 genotypes as indicated. Each lane represents an individual foetus. Note the absence of PHF6 protein at ~41 kDa in foetuses with germline deletion (Phf6–/Y;Nes-cre+/+) or nervous system-specific deletion (Phf6lox/Y;Nes-creTg/+) of the single allele of Phf6 in males or germline deletion of one allele and nervous system-specific deletion of the second allele in the female (Phf6–/lox;Nes-creTg/+). N = 8 individuals with BFLS, 1 BFLS carrier and 4 control individuals (A), 3 Phf6+/Y and 5 Phf6–/Y E15.5 mouse foetuses (B), 3 Phf6+/Y;Nes-creTg/+ and 4 Phf6lox/Y;Nes-creTg/+ E16.5 mouse foetuses (C) and 3 Phf6 deleted and 5 control E15.5 mouse foetuses, genotypes indicated above the blot (D). Data are displayed as mean ± sem and individual foetuses are represented by circles (B,C). RNA sequencing data (B,C) were analysed as described in the methods section; p values displayed above bars, two-way ANOVA with Šídák’s multiple comparisons test.