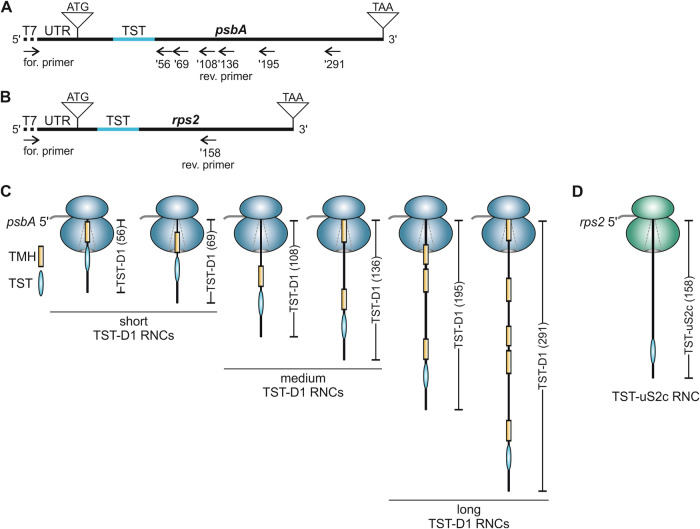
Figure 1. Generation of stalled affinity-tagged ribosome-nascent chain complexes (RNCs).
(A) Schematic representation of the psbA cDNA with T7 promotor sequence (dotted line) at the 5’ end and the endogenous psbA 5’ UTR (87 nt) used for PCR production of truncated templates for in vitro transcription into mRNA. A 90 nt Twin-Strep-tag (TST) coding sequence (blue line) was inserted at position +76 of the psbA sequence. The reverse primers (’56, ’69, ’108, ’136, ’195, and ‘291) lacking a stop codon, determine the length of the PCR product and are named according to the number of D1 amino acids encoded by the corresponding mRNA. (B) Scheme of the rps2 cDNA used for PCR production of a truncated template for in vitro transcription into mRNA (description as in (A)). The endogenous 5’ UTR comprises 90 nt. The TST coding sequence was inserted at position +52. The reverse primer (’158) lacks a stop codon and was used for PCR amplification of a product that corresponds to 158 amino acids of the uS2c protein. (C, D) Schematic RNCs generated by in vitro translation of mRNA from truncated templates as shown in (A) and (B). An internal TST is used for the affinity purification of the complexes. The nascent peptides of the D1 protein comprise at least one transmembrane helix (TMH) that is buried in the ribosome peptide tunnel or is exposed to the surrounding environment depending on the nascent peptide length. The nascent peptide of the soluble uS2c protein lacks any hydrophobic TMH.