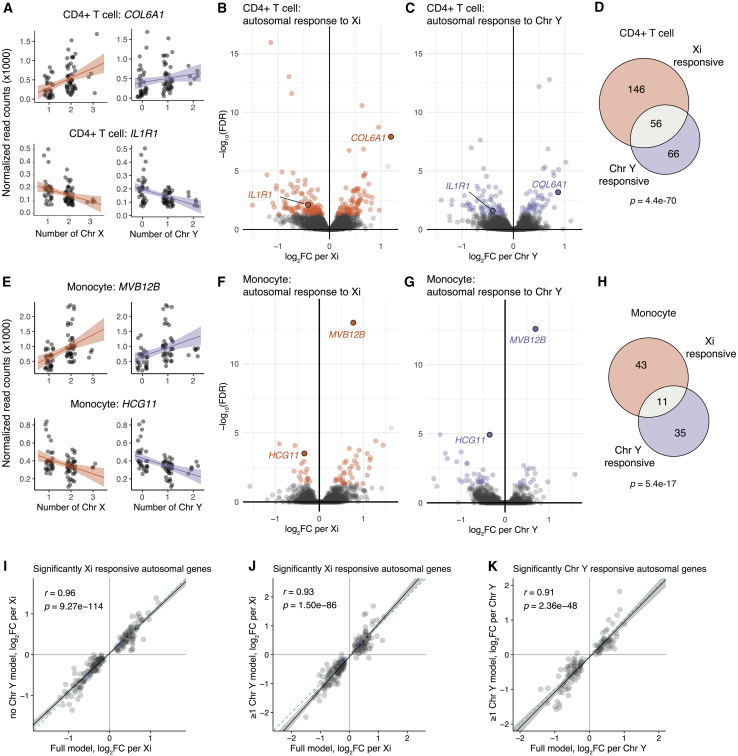
Figure 3.
CD4+ T cells and monocytes show significant autosomal gene responses to Xi and Chr Y dosage
(A) Examples of genes positively and negatively responsive to Xi (left) or Chr Y (right) dosage in CD4+ T cells. Regression lines with confidence intervals are shown.
(B and C) Volcano plots of autosomal responses in CD4+ T cells to Xi dosage (84 positively and 118 negatively responsive genes) (B) and Chr Y dosage (52 positively and 70 negatively responsive genes) (C).
(D) Overlap of autosomal genes with significant responses to Xi and Chr Y in CD4+ T cells; p value is from hypergeometric test.
(E) Examples of genes positively and negatively responsive to Xi (left) or Chr Y (right) dosage in monocytes.
(F and G) Volcano plots of autosomal responses in monocytes to Xi dosage (38 positively and 16 negatively responsive genes) (F) and Chr Y dosage (15 positively responsive and 31 negatively responsive genes) (G).
(H) Overlap of autosomal genes with significant responses to Xi and Chr Y dosage in monocytes; p value is from hypergeometric test.
(I–K) In CD4+ T cells, correlations of log2 fold-changes per Xi (using full model: all six karyotypes) against Xi or Chr Y dosage in subset models as indicated. Solid black line indicates Deming regression slope, with 95% confidence intervals shaded gray. Pearson’s correlation coefficients are shown; dashed blue lines indicate X = Y identity.