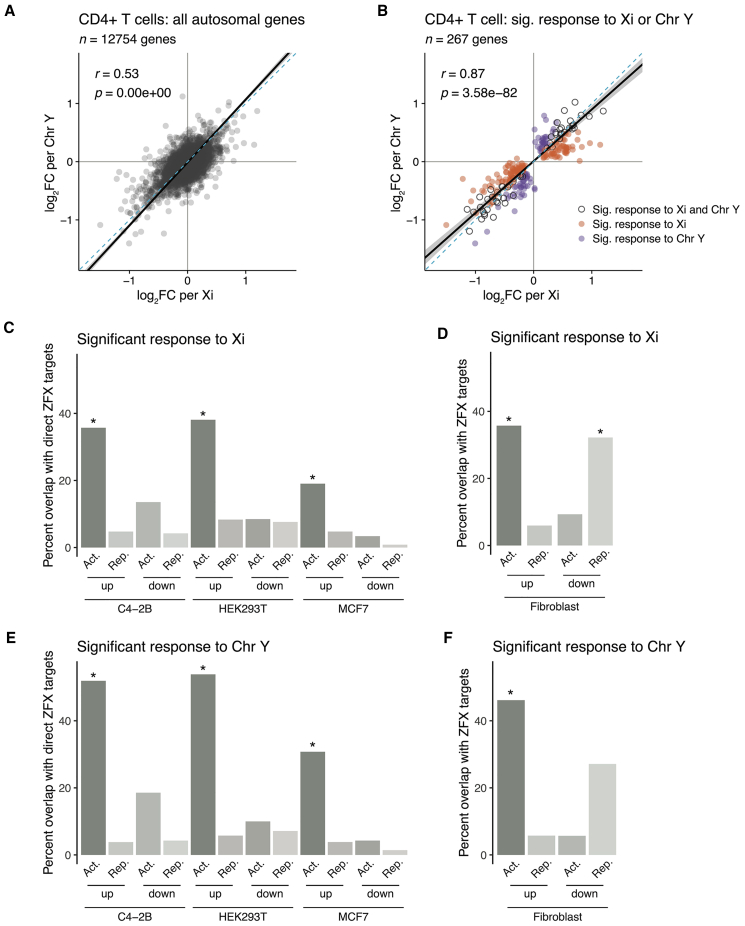
Figure 4.
The transcription factor ZFX drives a significant fraction of autosomal response to sex chromosome dosage in immune cells
(A and B) Scatterplot of log2 fold changes per Xi versus per Chr Y of all expressed autosomal genes in CD4+ T cells (A) or limited to genes significantly responsive to Xi dosage (orange), to Chr Y dosage (purple), or both (gray) (B). Solid black line indicates Deming regression slope, with 95% confidence intervals shaded gray. Pearson’s correlation coefficients are shown; dashed blue lines indicate X = Y identity.
(C–F) Bar plots showing percentages of genes with significant responses to Xi and/or Chr Y dosage in CD4+ T cells that were also identified as ZFX direct target genes in MCF7, C4-2B, and HEK-293T cell lines or ZFX responsive in fibroblasts. Genes are parsed by whether they significantly increased or decreased with Xi or Chr Y dosage (“up” or “down”) and were activated or repressed by ZFX (“Act.” or “Rep.”). p values reflect hypergeometric tests to identify significant enrichments of ZFX target genes in Xi- or Chr Y-responsive genes. Each comparison was restricted to genes expressed in both CD4+ T cells and the given in vitro cell line. Asterisks indicate the enrichments’ p values were lower than the Bonferroni-adjusted threshold of 0.001.