Figure 2.
Design and construction of the tumorized tumor-normal genome pairs
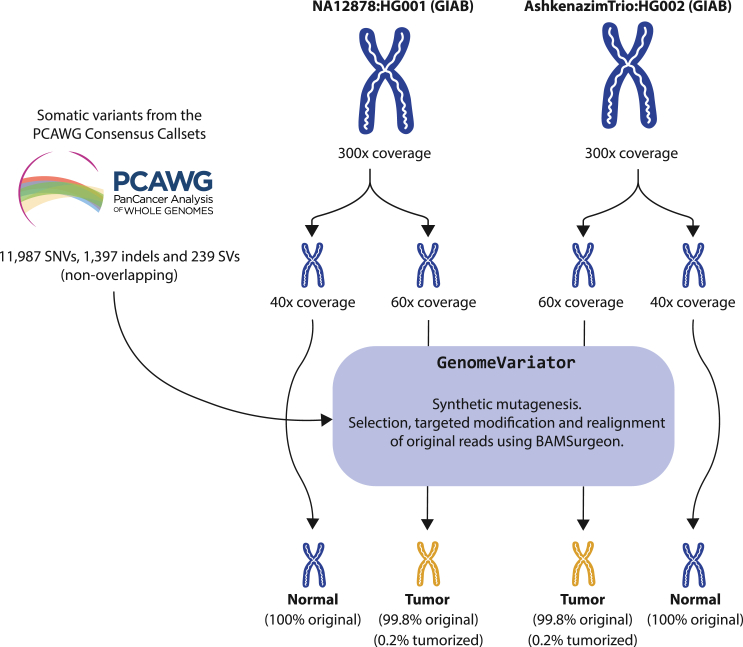
For an accurate assessment of the precision and to calculate the rate of false positives during somatic variant discovery, we have generated tumorized genomes. These consist of real WGS samples from the GIAB project43 with synthetic cancer somatic variants extracted from the PCAWG consensus callsets.4 For each introduced variant, a subset of reads in the tumor sample are modified to represent the variant. The number of modified reads depends on the depth of the region where the variant is located and the selected VAF (see Figure S6). This method is implemented in GenomeVariator, a wrapper tool that enhances the functionalities of BAMSurgeon.25,26,44 The high coverage of these samples (300×) allows the generation of tumor and normal genomes with a different composition of reads, recreating real tumor-normal analysis scenarios. Furthermore, the fact that only 0.2% of the reads have been modified to reconstruct the variants in the tumor samples of the tumorized genome pairs makes these samples ideal for an accurate evaluation of precision, as they retain 99.8% of the original sequencing and mapping properties. In order to avoid potential sample bias, we have generated two tumor-normal samples with the same validated variants: one derived from the NA12878 GIAB sample and the other from the HG002 GIAB sample.