Figure 1.
1000 Genomes Project and H7 human embryonic stem cell rRNA and rDNA variant extraction pipeline with high correlation between 28S rDNA and rRNA high-abundance variant frequencies
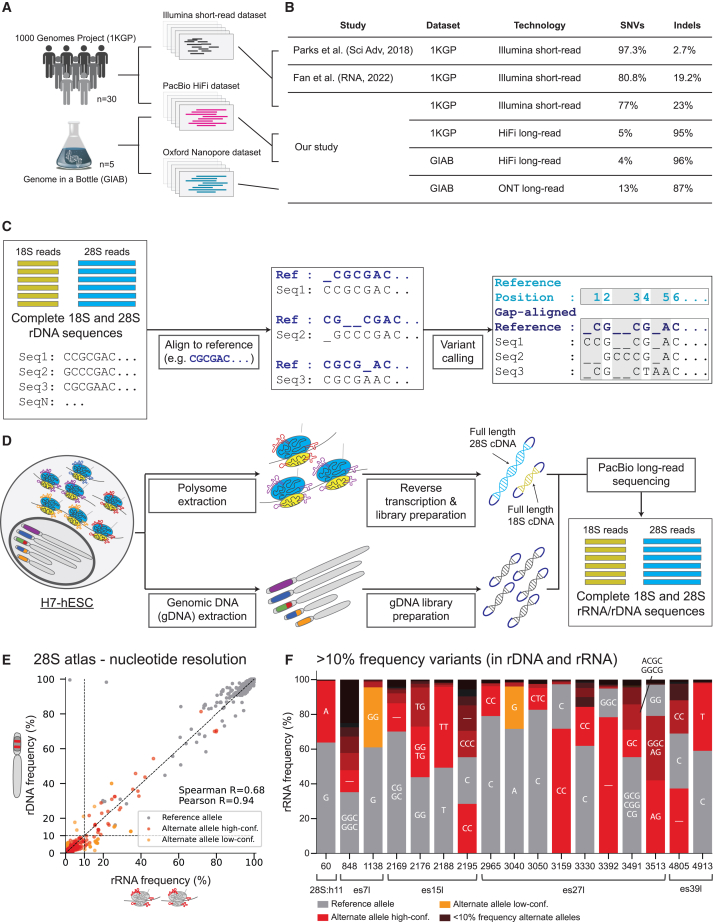
(A) Graphical illustration of the dataset analyzed consisting of 30 individuals from the 1000 genomes project (1KGP) with both short- and long-read sequencing.
(B) Comparison of single-nucleotide variants (SNVs) and insertion-deletions (indels) across studies.
(C) Graphical illustration of the reference gap alignment (RGA) method used for variant discovery in 18S and 28S sequences.
(D) 18S and 28S rDNA/rRNA sequence extraction pipeline from H7 human embryonic stem cell (H7-hESC).
(E) Scatterplot of 28S rRNA frequency (x axis) and rDNA frequency (y axis) for reference and alternate alleles. Alternate alleles are marked in red if their frequencies in rDNA and rRNA agree or in orange if they differ significantly. Reference alleles are marked in gray. A dashed black line indicates rRNA frequency equal to 10%. Spearman and Pearson correlations for rRNA frequency and rDNA frequency between alternate alleles alone are presented (calculated on variants, red dots alone).
(F) Stacked bar plots of allele frequencies at positions with variants with frequency >10% in both rRNA and rDNA. The nucleotide sequence matching the alleles are indicated inside the bar plots for variants with >10% allele frequency (“–” indicates deletion). The reference allele is indicated in gray, and alternate alleles are indicated in color.