Figure 4.
In-cell dimethyl sulfate with long-read sequencing shows that 28S subtypes have different RNA 2D structure
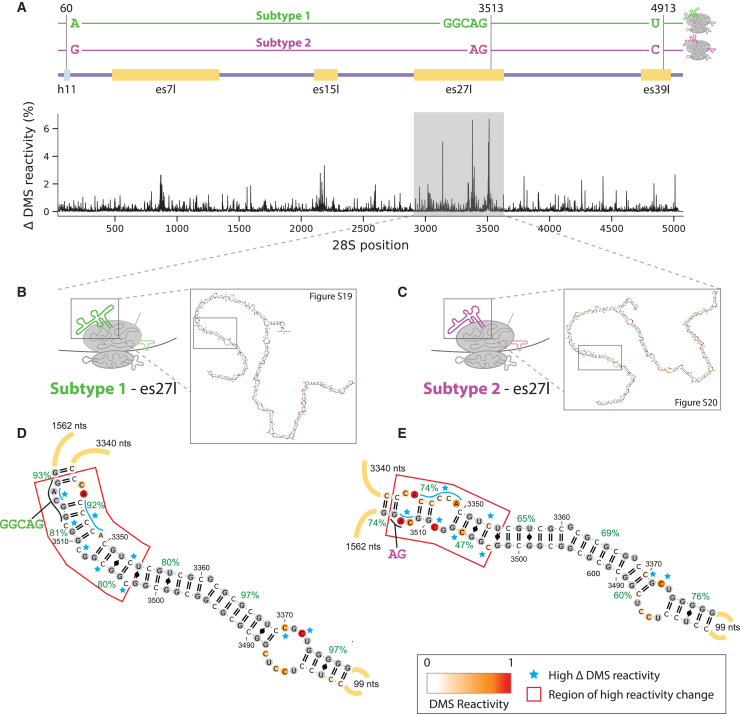
(A) Changes in dimethyl sulfate (DMS) accessibility between two most abundant 28S subtypes across the complete 28S molecule. Subtypes are defined by the sequence variants observed at positions 60 (28S:h11), 3513 (es27l), and 4913 (es39l), according to the numbering in NR_146117.1. Above is an illustration of the two subtypes, together with the annotations for the aforementioned regions and other regions with large differences in accessibility. x axis is the nucleotide position along the 28S, and y axis is the absolute percentage of DMS accessibility differences at a given position for a window size of 10 nucleotides.
(B) Illustration of es27l predicted secondary structure for subtype 1 (A,GGCAG,T). Detailed RNA 2D structure of the whole subtype 1 es27l is shown in Figure S19.
(C) Illustration of es27l predicted secondary structure for subtype 2 (G,AG,C). Detailed RNA 2D structure of the whole subtype 2 es27l is shown in Figure S20.
(D) Zoomed-in predicted RNA secondary structure of subtype 1 es27l between positions 3310 and 3552(+3). RNA secondary structures are colored by DMS reactivity, and helix confidence estimates are depicted as green percentages. Regions with major differences are annotated by the red box. Nucleotides with differing accessibility between the two subtypes are highlighted by blue stars.
(E) Zoomed-in predicted RNA secondary structure of subtype 2 es27l between positions 3310 and 3552. Detailed description of annotation is the same as (D).