Figure 4.
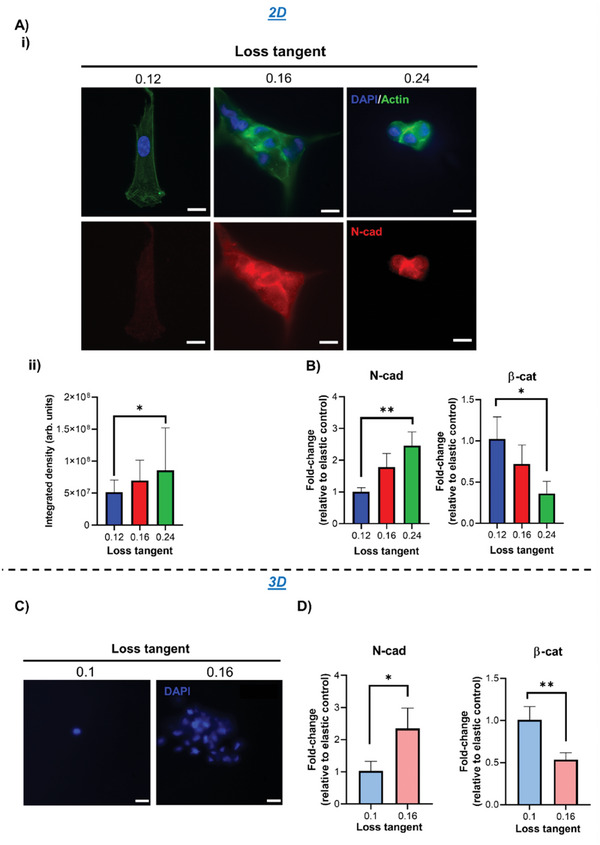
hMSC cell–cell signaling is enhanced in matrices with a higher loss tangent. A‐i) Representative immunofluorescence images of hMSCs cultured for 3 days on 2D PAAm hydrogels with DAPI (blue), actin (green), and N‐cadherin (red) staining, and ii) quantification of N‐cadherin expression by integrated density, n = 26. B) qPCR data from hMSCs cultured on 2D PAAm hydrogels for 3 days showing fold‐change in gene expression of N‐cadherin (left) and β‐catenin (right) relative to control with stronger elastic character (0.12 loss tangent samples) and normalized to GAPDH, n = 3. C) Representative images of DAPI‐stained hMSCs after 3 days of culture in 3D PEG‐MAL hydrogels. D) qPCR data from hMSCs cultured in 3D PEG‐MAL hydrogels for 3 days showing fold‐change in gene expression of N‐cadherin (left) and β‐catenin (right) relative to the control with stronger elastic character (0.1 loss tangent samples) and normalized to GAPDH, n = 3. For all figures, data are represented as mean ± standard deviation, and differences are considered significant for p ≤ 0.05 using one‐way ANOVAs and t‐test for multiple and pairwise comparisons respectively (* p ≤ 0.05, ** p ≤ 0.01). Integrated density quantification was normalized to cell number based on the number of nuclei in each image. All hydrogels were functionalized with 2 mm RGD peptide to allow cell adhesion. All PEG‐MAL hydrogels were crosslinked using peptide ratios of 1% VPM and 99% scrambled VPM. Scale bars = 20 µm.
