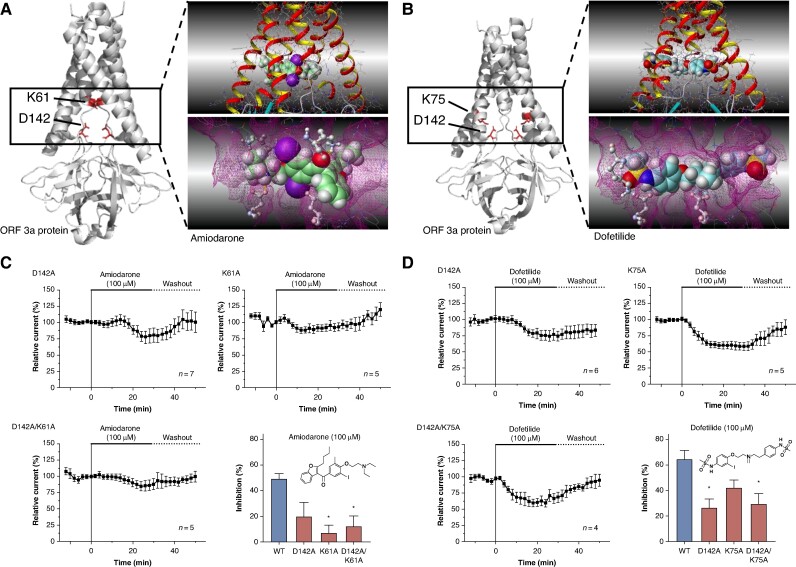
Figure 5.
Structural determinants of SARS-CoV-2 ORF 3a protein interaction with amiodarone and dofetilide. (A and B) Computational docking simulation revealing the interaction of amiodarone (A) and dofetilide (B) with the intracellular channel pore of the SARS-CoV-2 ORF 3a protein dimer. The simulation is based on the cryo-EM structure delineated by Kern et al.22 (PDB-ID: 7KJR). It highlights the critical roles of amino acids lysine 61 (K61) and aspartic acid 142 (D142) in forming the molecular drug-binding site for amiodarone (A) and the involvement of lysine 75 (K75) along with aspartic acid 142 (D142) in the interaction site for dofetilide (B). (C) To confirm the contribution of the said amino acids to the drug-binding site, mutant proteins SARS-CoV-2 ORF 3a K61A, D142A, and the double-mutant K61A/D142A were expressed in X. laevis oocytes and exposed to amiodarone (100 µM; 30 min as described above). (D) In a similar fashion, SARS-CoV-2 ORF 3a-K75A, D142A, and the double-mutant K75A/D142A were superfused with dofetilide (100 µM; 30 min as described above) upon heterologous expression in X. laevis oocytes. Data are presented as mean ± standard error of the mean. Chemical structures of amiodarone and dofetilide are depicted as insets. Statistical significance is indicated by *, representing a P < 0.05 from a two-tailed unpaired Student’s t-test vs. WT channels followed by Bonferroni correction.