Abstract
GLI proteins are involved in the development of mice, humans, zebrafish, Caenorhabditis elegans, Xenopus, and Drosophila. While these zinc finger-containing proteins bind to TG-rich promoter elements and are known to regulate gene expression in C. elegans and Drosophila, mechanistic understanding of how regulation is mediated through naturally occurring transcriptional promoters is lacking. One isoform of human GLI-2 appears to be identical to a factor previously called Tax helper protein (THP), thus named due to its ability to interact with a TG-rich element in the human T-lymphotropic virus type 1 (HTLV-1) enhancer thought to mediate transcriptional stimulation by the Tax protein of HTLV-1. We now demonstrate that, working through its TG-rich binding site and adjacent elements, GLI-2/THP actually suppresses gene expression driven by the HTLV-1 promoter. GLI-2/THP has no effect on the HTLV-2 promoter, activates expression from the promoters of human immunodeficiency virus types 1 and (HIV-1 and -2), and stimulates HIV-1 replication. Both effective suppression and activation of gene expression and viral replication require the first of the five zinc fingers, which is not necessary for DNA binding, to be intact. Thus, not only can GLI-2/THP either activate or suppress gene expression, depending on the promoter, but the same domain (first zinc finger) mediates both effects. These findings suggest a role for GLI-2 in retroviral gene regulation and shed further light on the mechanisms by which GLI proteins regulate naturally occurring promoters.
Human retroviruses have usurped cellular signal transduction pathways and proteins to regulate their transcription and hence replication. These cellular proteins, which often interact with retroviral promoters, are frequently members of proto-oncogene families and are similar to proteins involved in the regulation of development. Different sets of cellular proteins are involved in regulating the transcription of the four known human retroviruses: human T-lymphotropic virus type 1 (HTLV-1), which is known to cause leukemias and lymphomas and to be involved in the pathogenesis of tropical spastic paraparesis (29, 62, 66, 67, 79, 94); HTLV-2 (43), which is not yet definitively linked with any human disease; and human immunodeficiency virus types 1 and 2 (HIV-1 and -2), which are known to cause AIDS (5, 15–17, 27, 34, 68). Each of these viruses makes use of a specific set of cellular proteins to activate transcription, often in response to T-cell stimulation. (Fig. 1 and references 13, 24, 25, 36, 37, 39, 46, 48, 54, 55, 58, and 64).
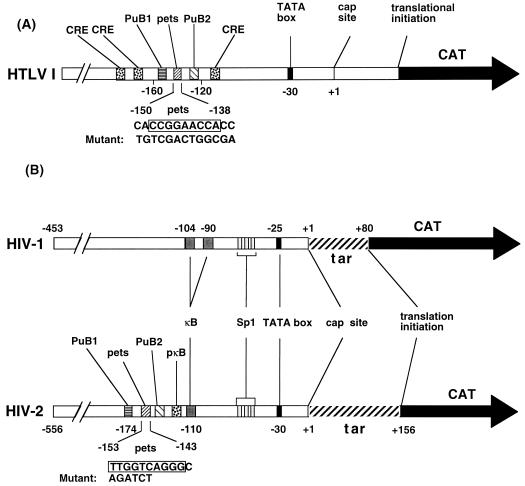
FIG. 1.
Enhancer regions of HTLV-1 (A) and HIV-1 and HIV-2 (B). Relevant sites within the LTR sequences are identified. The altered bases within the mutant pets plasmids used are shown below the wild-type sequence. The sequence of HIV-2ROD has been published elsewhere (33). The sequence of the pets site of the HTLV-1 enhancer differs slightly from the previously published sequence in which the G at position −144 on the coding strand is shown as AA (8, 28). CRE, CREB response element (=TRE-1); tar, Tat activation response element. PuB1 and PuB2 bind Ets family members (14, 30, 48, 55). The pets site is discussed in the text. The HIV-2 κB and peri-κB (pκB) sites have been described previously (13, 54), as have the HIV-1 κB and Sp1 sites (58, 59). The putative HIV-2 Sp1 sites have not been tested experimentally, other than as shown in Fig. 7.
Within the HTLV-1 enhancer, there are two types of elements which are thought to mediate transcriptional activation in response to the viral Tax protein, known as Tax-responsive elements 1 and 2 (TRE-1 and -2) (30, 41, 60, 69, 82). The TRE-1 motifs consist of 21-bp repeats which bind the cyclic AMP response element binding protein (CREB). Interestingly, within the TRE-2 region, there is a TG-rich element, similar to the HIV-2 peri-ets (pets) site, which we have shown to be important to the response of both promoters to T-cell activation (14, 24, 25, 36, 37, 39, 55). While other groups have suggested that this region is Tax responsive (85, 86), our results did not support that contention (14). Yoshida and colleagues used Southwestern screening to clone a protein which binds to the TRE-2 pets-like element. This protein, termed Tax helper protein (THP), has been found in two splice variants, THP-1 and THP-2, which differ only in that THP-2 has an extra 17 amino acids (aa) near the amino terminus (86). Subsequent analysis has shown that THP, a protein with five zinc fingers, is a form of human GLI-2 and that multiple isoforms of GLI-2 exist, of which THP is the smallest (85). The human GLI proteins are closely related to the Caenorhabditis elegans sex determination protein tra-1 (61, 95, 96) and less closely related to Krüppel proteins (50, 97) and to YY1, a zinc finger-containing protein known for its ability to stimulate or repress transcription (51, 80, 87). The Drosophila protein cubitus interruptus is a GLI protein which is known to regulate gene expression and to be important in fly development. Recently, GLI proteins have also been found in Xenopus (53) and in zebrafish (45). GLI proteins play an integral part in the Hedgehog signaling pathway, which is involved in normal and abnormal cellular development (12, 35, 52, 84).
There are three known human GLI proteins: GLI-1, GLI-2, and GLI-3. GLI-1, the prototypical GLI protein, can transform cells in cooperation with adenovirus E1A and has been identified as being amplified in certain human gliomas (hence the name) and in certain human sarcomas (47, 73, 76, 77; but also see reference 91) and has been linked to basal cell carcinomas (18). GLI-3 has been shown to be the protein mutated in human Greig's syndrome (and in the mouse equivalent), in which facial and limb abnormalities are seen (89), in Pallister-Hall syndrome (44), and in postaxial polydactyly type A (71). The wild-type and mutant forms of GLI-3 bear some localization and functional similarities to the longer (activation) and shorter (repression) forms of cubitus interruptus (81). Like GLI-1 and GLI-3, GLI-2 is expressed at high levels in glioblastoma multiforme (76). It has also been shown that GLI-2 mutant mice have diminished Sonic Hedgehog signaling and severe skeletal abnormalities including cleft palate, tooth defects, absence of vertebral body and intervertebral discs, and shortened limbs and sternum (22, 38, 56). Mice mutant in both GLI-2 and GLI-3 also show abnormal development of the lung, trachea, and esophagus (57). While Krüppel and cubitus interruptus are known to modulate gene expression, and while other human GLI proteins are known to bind to TG-rich elements similar to the site to which THP (GLI-2) binds, little has been known about the effect of human GLI proteins on gene expression or about naturally occurring promoters which might respond to these proteins. Further, while multiple copies of GLI binding sites placed upstream of a heterologous promoter can be activated or suppressed by GLI proteins (1, 53, 78), it has not generally been ascertained whether the TG-rich GLI binding elements mediate the effects that GLI proteins might have on transcription driven by natural promoters.
In view of the fact that GLI-2/THP is capable of binding to an important regulatory element of the HTLV-1 promoter, and in view of the limited information available on the effect of human GLI proteins on gene expression, we tested the effect of cotransfecting GLI-2/THP with the HTLV-1 promoter. We now demonstrate that contrary to what might be expected from a putative Tax helper protein, GLI-2/THP actually decreases expression from the HTLV-1 promoter but has no effect on the HTLV-2 promoter. Mutation of a single amino acid in the first zinc finger, which is not necessary for DNA binding, markedly affects the ability of GLI-2/THP to modulate expression from the HTLV-1 promoter. In contrast to the effect on HTLV-1, GLI-2/THP activates HIV-1 replication and increases expression from both the HIV-2 and HIV-1 promoters, an effect which is also dependent on the first and second zinc fingers and is reflected at the RNA level. The pets site and surrounding enhancer elements mediate the response of HTLV-1 to GLI-2/THP, but surprisingly, neither the pets site nor other upstream enhancer elements mediate its effect on HIV. Last, we show that only THP, the truncated form of human GLI-2, significantly modulates retroviral transcription. Thus, these studies favor the interpretation that human GLI proteins, like cubitus interruptus, serve both transcriptional activation and repression functions, but unlike the case for cubitus interruptus, the same form of the protein can serve both purposes.
MATERIALS AND METHODS
Cell culture and transfections.
The CV-1 monkey kidney cell line was cultured in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum, 2 mM l-Glutamine, and penicillin-streptomycin. Cells were plated at 50% confluency, transfected by the calcium phosphate method, shocked with 10% dimethyl sulfoxide in phosphate-buffered saline after 4 h, and harvested for chloramphenicol acetyltransferase (CAT) assays 40 h posttransfection. The Jurkat T-cell line, the U937 monocytic cell line, and the U1 HIV-1-infected monocytic cell line were grown in RPMI 1640 supplemented with 10% fetal bovine serum, 2 mM l-glutamine, and penicillin-streptomycin. Cells (107) were transfected by the DEAE-dextran method (70), stimulated where indicated with 16 nM phorbol myristate acetate (PMA) after 20 h, and harvested after an additional 20 h of incubation. In all transfections, cell lysates were prepared by multiple freeze-thaw cycles in 0.25 M Tris-Cl (pH 7.5), and CAT activity was assayed by standard methods (31). Transfection efficiencies were normalized for protein concentration or Rous sarcoma virus promoter-driven luciferase expression, measured using the Bio-Rad reagent or Promega Genelight and a Wallac scintillation counter, respectively. CAT activity was quantitated on a Betagen beta scanner.
Plasmids.
The HTLV-1, HTLV-2, HIV-1, and HIV-2 long terminal repeat (LTR)-CAT reporter constructs have been described elsewhere (30, 54, 83), as have the HTLV-1 Δpets, HIV-2 Δpets, HIV-2 ΔκB, and HIV-2 −80 truncation (14, 54, 55). Plasmids containing the HIV-1 mutation ΔκB or ΔTATA (6, 58) or a depletion of all three Sp1 sites (59) were gifts from Gary Nabel. HIV-2 ΔSp1, in which the 3′ putative Sp1 site is mutated, was created using an Altered Sites II (Promega) site-directed mutagenesis kit to convert the HIV-2 wild-type sequence from 5′-GGGAGGAGCTGGTGGGGAACGCCC-3′ to the mutant sequence (underlined) 5′-GGGAGGAGCGGATCCGGAACGCCC-3′. The sequence was confirmed by dideoxynucleotide sequencing with a Sequenase kit (Amersham). The deletion mutants of the β form of human GLI-2 were made with an Exo-Size deletion kit from New England Biolabs according to the protocol provided. The XbaI-to-BamHI GLI-2 β insert was ligated into pGEM 7f− for deletion mutagenesis, and BamHI linkers were ligated to the exonuclease III-digested ends. The XbaI-BamHI-digested fragments were isolated by agarose gel electrophoresis and ligated back into the original cytomegalovirus (CMV)-based pCG expression vector. Clones of interest were sequenced by the University of Michigan automated sequencing core. HTLV-1 and HTLV-2 Tax expression plasmids were previously described (10, 30, 49). The GLI-2/THP expression plasmids pCG-THP-1 and pCG-THP-2 were constructed and kindly provided by M. Yoshida (86), and mutations of the first zinc finger were introduced using an Altered Sites kit. Point mutations (C to G) were made in either one (ΔCys 1; also referred to as GLI mutant finger [GLI mtF1] for simplicity in Fig. 10]) or both (ΔCys 1 + 2) cysteine-encoding sequences of the first zinc finger, thus converting cysteine codons to tryptophan codons. The same methodology was used to create constructs in which the first cysteine in the second zinc finger (GLI mtF2) or the first cysteine in both zinc fingers (GLI mtF1F2) were mutated. The GLI-2/THP–glutathione S-transferase bacterial fusion protein constructs were made by using PCR to add in-frame BamHI sites to the ends of the GLI-2/THP coding sequence and cloning the full-length THP-2 BamHI or a 3′-truncated BamHI-to-SmaI fragment into pGEX-2TK (Pharmacia).
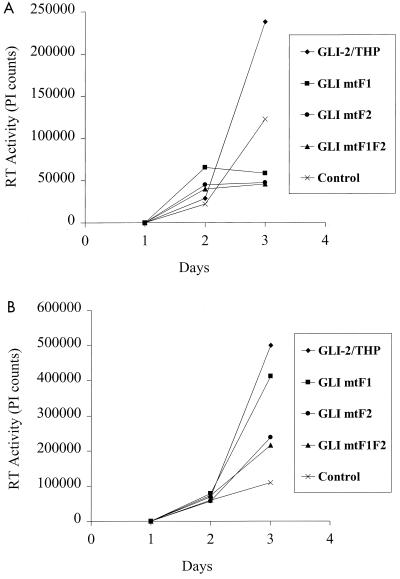
FIG. 10.
GLI-2/THP stimulation of HIV-1 replication is mediated by the first and second zinc fingers. (A) U937 monocytic cells were transfected with 2 μg of the infectious HIV-1 clone p89.6 and 2.5 μg of GLI-2/THP, GLI mtF1, GLI mtF2, GLI mtF1F2, or control vector as indicated. The cells were treated with PMA at a concentration of 16 nM 24 h after transfection. RT activity was measured in supernatants collected each day after transfection. The data represent mean RT activity in triplicate wells and are representative of four separate experiments. (B) The chronically infected monocytic cell line U1 was transfected with 5 μg of GLI-2/THP, GLI mtF1, GLI mtF2, GLI mtF1F2, or control vector as indicated. The cells were treated with PMA at a concentration of 16 nM 24 h after transfection. RT activity was measured in supernatants collected each day after transfection. The data represent mean RT activity in triplicate wells and are representative of two separate experiments.
Primer extension and RNase protection assays.
Total cellular RNA from transfected U937 cells was isolated by a modification of the method of Chomczynski and Sacchi (9, 39). Primer extensions were performed as described in the Promega protocol. A primer complementary to the 5′ end of the CAT gene (5′-TGCCATTGGGATATATCAACGGTG-3′) was 5′ end labeled with 32P and hybridized for 30 min at 58°C with 10 μg of RNA in 10 μl of avian myeloblastosis virus (AMV) reverse transcriptase (RT) buffer. To the hybridization mixture was added 10 μl of 1× AMV RT reaction buffer containing 1.4 μl of 40 mM sodium pyrophosphate and 1 U of AMV RT (Promega), and the reaction mixture was incubated at 42°C for 1 h. The primer extension products were then precipitated and analyzed by electrophoresis through a 6% polyacrylamide gel containing 7 M urea. RNase protection assays were performed as described elsewhere (9). The GLI-2/THP probe was prepared using the wild-type GLI-2/THP cDNA cloned into pcDNA3.1/Neo (Invitrogen) linearized with BstEII and transcribed by SP6 polymerase with the Riboprobe system (Promega) and [α-32P]CTP as label. The CAT probe was made by inserting the EcoRI fragment of SP-CAT into pGEM 7f− (Promega) and transcribing with T7 polymerase. RNA (5 μg) from transfected cells was hybridized at 48°C overnight with 105 cpm of the appropriate probe and digested with RNase T1 for 1 h at 30°C. The RNA was then precipitated with an equal volume of 4 M guanidinium thiocyanate, 10 μg of tRNA carrier, and 2 volumes of isopropanol and analyzed on a 6% polyacrylamide–7 M urea gel. The protected fragments were quantitated on a PhosphorImager (Molecular Dynamics).
EMSAs with recombinant GLI-2/THP and deletion mutants.
GLI-2/THP and mutant GLI-2/THP were first prepared as described in the accompanying report (9a). The recombinant proteins were released from the agarose beads by digestion with thrombin. Protein was quantitated by a commercially available kit (Bio-Rad). Five micrograms of recombinant protein was incubated at room temperature with the HIV-2 pets probe in the presence or absence of 100 ng of competitor. Electrophoretic mobility shift assays (EMSAs) were performed as described previously (39, 55). The reaction products were analyzed on a nondenaturing 5% polyacrylamide gel in 1× Tris-borate-EDTA buffer.
HIV-1 replication studies.
HIV-1 replication in U937 monocytic cells was assessed by transfecting plasmids encoding GLI-2/THP, GLI mutants, or control vector along with a biologically active infectious molecular clone of the dual-tropic cytopathic HIV-1 primary isolate 89.6 (p89.6) by the DEAE-dextran method. Viral replication was assessed for 3 days following transfection by RT assay as described elsewhere (2, 70). HIV-1 replication in the chronically infected U1 monocytic cells was also assessed by RT assay following transfection with the GLI constructs by the DEAE-dextran method.
RESULTS
GLI-2/THP decreases expression from the HTLV-1 but not HTLV-2 promoter.
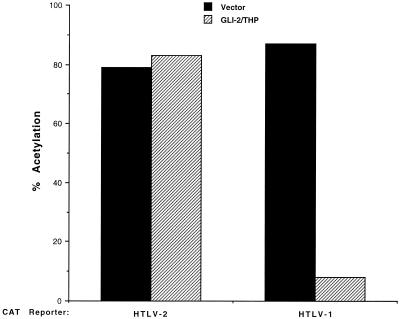
In view of the ability of GLI-2/THP to bind to the pets-like site in the HTLV-1 promoter (Fig. 1A), we tested whether this protein could modulate HTLV-1 and HTLV-2 gene expression in contransfection assays. An expression vector encoding GLI-2/THP or an empty control vector was transfected along with a construct expressing CAT under the control of the HTLV-1 or HTLV-2 promoter. Contrary to expectations, GLI-2/THP actually decreased expression from the HTLV-1 promoter (Fig. 2). This effect was dose responsive and was seen with the lowest amount of GLI-2/THP (2 μg) tested (data not shown). In contrast, GLI-2/THP had no effect on the HTLV-2 promoter (Fig. 2). The suppressive effect of GLI-2/THP on HTLV-1 was present whether the promoter-driven expression was tested at basal levels (Fig. 3) or when the promoter was stimulated with Tax (Fig. 2). Therefore, we conclude that GLI-2/THP can suppress HTLV-1 expression but appears to have no effect on the related HTLV-2 promoter.
FIG. 2.
GLI-2/THP suppresses expression from the HTLV-1 but not the HTLV-2 LTR. CV-1 cells were cotransfected with 5 μg of HTLV-1 or HTLV-2/CAT, 0.5 μg of the respective Tax expression plasmid, and 7.5 μg of either CMV-driven GLI-2/THP (pCG-THP) or the empty vector. Cell extracts were prepared after 48 h, normalized for protein concentration, and assayed for CAT activity.
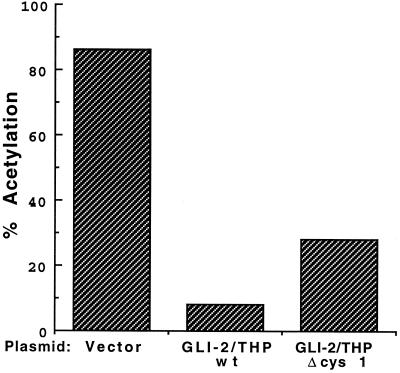
FIG. 3.
The first zinc finger contributes to suppression of the HTLV-1 LTR by GLI-2/THP. CV-1 cells were transfected with 5 μg of HTLV-1/CAT and 7.5 μg of the GLI-2/THP wild type (wt), GLI-2/THP ΔCyst 1 (in which the first zinc finger of GLI-2/THP has been disrupted), or control CMV promoter-containing vector. Cellular extracts were prepared 48 h later, normalized for protein concentration, and assayed for CAT activity.
The first zinc finger of GLI-2/THP mediates much of the effect on HTLV-1.
While the crystal structure of GLI-2/THP is not known, the structure of the prototypical GLI protein, GLI-1, has been analyzed (63). This study demonstrated that of the five zinc fingers, fingers 3, 4, and 5 closely contact DNA. Finger 2 partially contacts DNA, and finger 1 does not contact DNA. Therefore, we speculated that finger 1 might be involved in protein-protein interactions and hence might be involved in the modulation of gene expression. Thus, a single cysteine residue was mutated in the first zinc finger of GLI-2/THP. The mutated construct still made a full-length protein (not shown). Further, mutant protein was still able to bind to the HTLV-1 pets site (not shown). However, when transfected with the HTLV-1 promoter, the construct with the mutated zinc finger had a much less marked effect on HTLV-1 gene expression than did the wild type (Fig. 3). Therefore, we conclude that the first zinc finger is important to the function of GLI-2/THP.
The decrease in expression from HTLV-1 mediated by GLI-2/THP requires intact upstream enhancer elements which include the pets site.
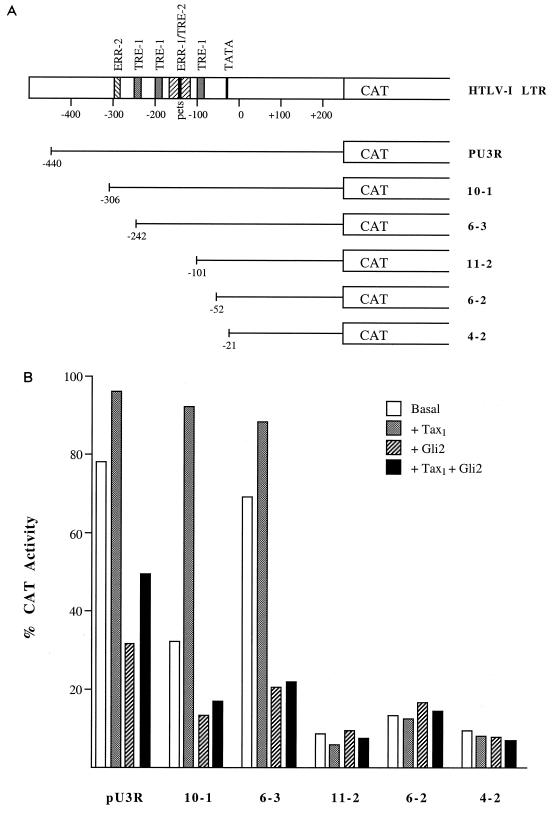
In view of the above information, we reasoned that the suppression of the HTLV-1 promoter by GLI-2/THP was mediated by the pets-like site in HTLV-1 (Fig. 1). We therefore cotransfected GLI-2/THP with the HTLV-1 wild-type promoter or the HTLV-1 promoter in which the pets site had been altered by site-specific mutagenesis (14). Surprisingly, although mutation of this site decreased basal promoter-driven expression, as we have seen previously (14), it did not alter the response to GLI-2/THP (not shown). Therefore, we tested a series of previously constructed deletion mutants (Fig. 4A) to assess which sites in the HTLV-1 promoter mediate the response to GLI-2/THP. Interestingly, when the region between −242 and −101 was deleted, both Tax responsiveness and GLI-2/THP suppression were lost (Fig. 4B). This region contains two of the 21-bp TRE-1s and the TRE-2, the latter of which contains the pets site. Thus, while the minimal pets site alone does not mediate the GLI-2/THP response, it is part of an enhancer complex which does.
FIG. 4.
The pets site and adjacent enhancer elements mediate the response of the HTLV-1 promoter to GLI-2/THP. (A) HTLV-1 LTR-CAT reporter plasmids. Full-length (pU3R) and 5′ deletion mutant HTLV-1 LTR sequences were cloned upstream of a CAT reporter gene for use in transient transfection assays in CV-1 cells as previously reported (8, 30). The HTLV-1 TRE-1 and TRE-2, the Ets-responsive regions (ERR-1 and ERR-2), the TATA box, and the transcription start site (nucleotide 0) are indicated. The TRE-2 element includes the pets site. Illustrated are the HTLV-1 sequences included upstream of the CAT reporter gene for the various LTR constructs and the regulatory domains that remain. (B) Effects of GLI-2/THP on basal and HTLV-1 Tax-stimulated transcriptional activity of the HTLV-1 LTR. Full-length and mutated HTLV-1 LTR-CAT reporter plasmids (7.5 μg) were transiently transfected into CV-1 cells (30) alone (basal) or with an expression plasmid for Tax1 (0.5 μg), GLI-2/THP (5 μg), or both. Cells were harvested 48 h after transfection, and CAT assays were performed with 10 μg of each extract. Shown are the percentages of acetylation of [14C]chloramphenicol (percent CAT activation) observed in a representative experiment.
GLI-2/THP requires an intact first zinc finger to increase expression from the HIV-1 and HIV-2 promoters.
We next asked whether GLI-2/THP can modulate the expression of either HIV-2, which contains a pets site (Fig. 1B), or HIV-1, which does not. As can be seen in Fig. 5A, both wild-type GLI-2/THP (lane 1) and GLI-2/THP in which the first zinc finger is mutated (lane 4) were able to bind to the HIV-2 pets site in vitro. Further, GLI-2/THP increased expression from the HIV-1 (see Fig. 7A) and HIV-2 (Fig. 5B; see also Fig. 7B) promoters, but only in the presence of PMA. RNase protection assays demonstrated that this was not a result of increased expression of GLI-2/THP with PMA treatment (Fig. 6B, lane 3 versus lane 4 and lane 7 versus lane 8). Here again, GLI-2/THP constructs with mutations in the first or first and second cysteines of the first zinc finger largely lost the ability to modulate gene expression (Fig. 5B). We therefore conclude that in the presence of PMA, GLI-2/THP can increase expression from HIV promoters, in contrast to its ability to depress expression of HTLV-1. Just as it is necessary for suppression of the HTLV-1 promoter, the first zinc finger is needed for activation of the HIV promoters.
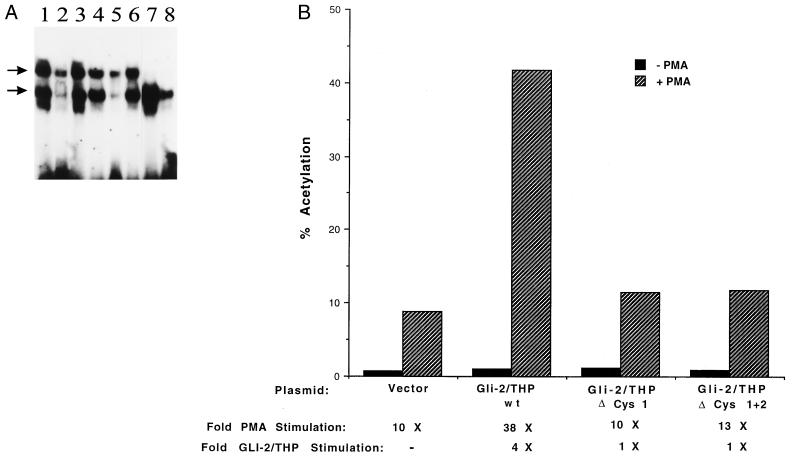
FIG. 5.
(A) GLI-2/THP and GLI-2/THP mutant in the first zinc finger bind to the HIV-2 pets site. The HIV-2 pets site probe was incubated with 5 μg of purified recombinant (made in Escherichia coli) GLI-2/THP (lanes 1 to 3) or mutant GLI-2/THP (lanes 4 to 6) without competitor (lanes 1 and 4), with 100 ng of unlabeled HIV-2 pets oligonucleotide (lanes 2 and 5), or with 100 ng of unlabeled mutated pets oligonucleotide (lanes 3 and 6). As a control for bacterial pets binding proteins, the HIV-2 pets site was also incubated with 5 μg of control extract which was prepared by isolating protein from E. coli programmed with the empty pGEX vector (lanes 7 and 8). Lane 7 represents incubation of the control extract in the absence of competitor and lane 8 in the presence of 100 ng of HIV-2 pets oligonucleotide. The upper arrow marks the mobility of the GLI-2/THP complex, and the lower arrow marks the mobility of a bacterial pets binding protein, which we have previously demonstrated to be the Lon protease (26). (B) GLI-2/THP increases expression from the HIV-2 LTR; this effect is dependent on PMA stimulation and can be greatly reduced by mutation of the first zinc finger. Jurkat cells were transfected with HIV-2 CAT and either the empty vector or the wild-type or mutant GLI-2/THP expression plasmid. The ΔCys 1 and ΔCys 2 mutations both disrupt the first zinc finger. Cell extracts were prepared at 48 h and normalized for protein concentration, and CAT assays were performed.
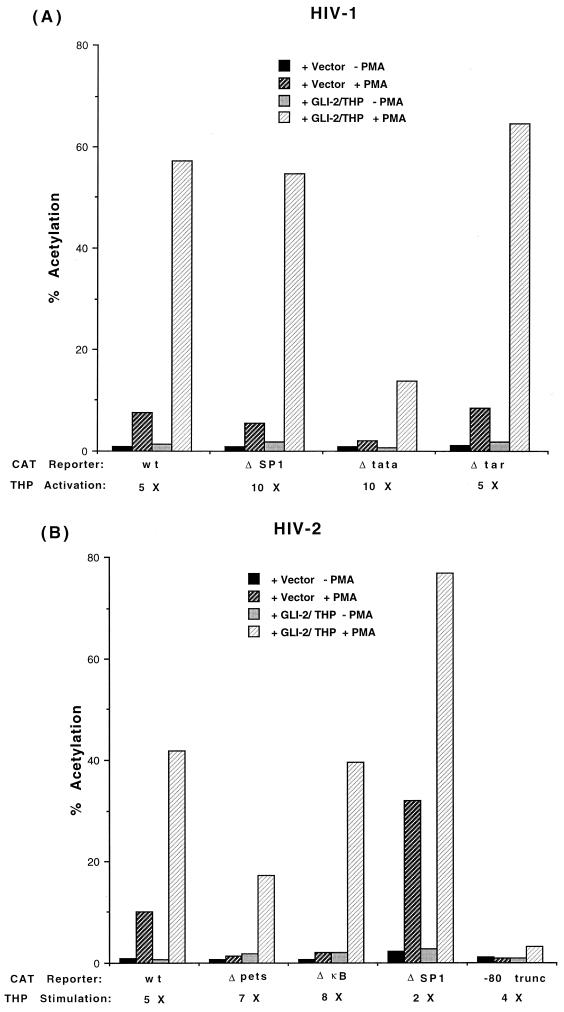
FIG. 7.
The effect of GLI-2/THP on HIV-1 and HIV-2 expression is not dependent on previously defined promoter elements. (A) Mutation of the κB or TATA sites, or deletion of the Sp1 sites or TAR element of HIV-1, has no effect on GLI-2/THP activation. (B) Mutation of the pets, κB, or 3′ Sp1 site or deletion of all cis-acting elements 5′ of position −80 in the HIV-2 promoter does not significantly diminish the effect seen with GLI-2/THP overexpression. The experiments shown were performed in Jurkat T cells. wt, wild type.
FIG. 6.
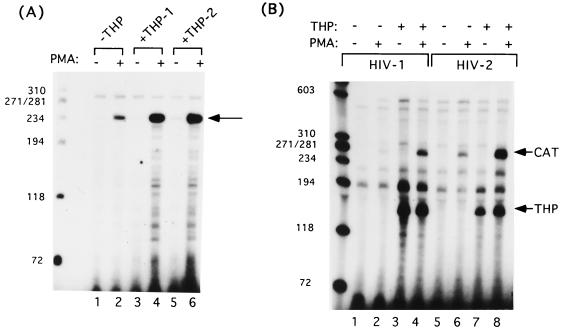
The GLI-2/THP-mediated increase in HIV LTR-driven CAT expression is reflected at the RNA level. (A) Primer extension analysis of RNA from GLI-2/THP- and HIV-2/CAT-cotransfected U937 cells. THP-1 and THP-2, splice variants of GLI-2/THP, both increase the level of CAT transcript approximately 10-fold in the presence of PMA. The arrow indicates the correctly initiated CAT transcript. (B) RNase protection assay of RNA from U937 cells cotransfected with either HIV-1 or HIV-2/CAT and GLI-2/THP. GLI-2/THP expression increases the level of CAT transcripts in PMA-stimulated U937 cells, while the level of GLI-2/THP message is not affected. Sizes are indicated in base pairs.
The increase in expression from the HIV-2 promoter driven by GLI-2/THP is reflected at the RNA level.
As the pets site did not mediate the effect of GLI-2/THP on HIV expression (Fig. 7B), we wished to ascertain that the effect was indeed seen at the RNA level. Therefore, we cotransfected HIV-2 and HIV-1 with GLI-2/THP in the presence of PMA and assessed the effect of adding GLI-2/THP plus PMA by primer extension assays. As shown in Fig. 6A, increases in RNA equivalent to the increases in CAT activity were seen (lane 3 versus lane 4, lane 5 versus lane 6, and lanes 4 and 6 versus lane 2). This was confirmed using RNase protection assays (Fig. 6B, CAT message in lane 2 versus lane 4 and lane 6 versus lane 8). Therefore, we conclude that the GLI-2/THP effect on retroviral gene expression is mediated at the RNA level.
The effect of GLI-2/THP on HIV-1 and HIV-2 gene expression is not mediated by upstream enhancer elements or the Sp1 or TATA sites.
The effect of GLI-2/THP on HIV gene expression was seen at the RNA level (Fig. 6) but surprisingly was not mediated by the pets site (Fig. 7B). We assessed whether other known regulatory regions of either HIV-2 or HIV-1 could be demonstrated to be responsible for this effect. As shown in Fig. 7B, even the deletion of all upstream enhancer elements of HIV-2 (the −80 truncation mutant), while markedly diminishing the response to PMA alone as previously reported (54, 55), did not affect the GLI-2/THP-stimulated increase in HIV-2 gene expression. Testing of the Sp1, TATA, and κB elements using HIV-1 constructs (Fig. 7A) demonstrated that these promoter elements were also not necessary for the GLI-2/THP effect to occur. tra-1, the GLI protein of C. elegans, has recently been shown to bind to RNA (32). While in C. elegans this binding affected posttranscriptional events, this observation did suggest that GLI-2/THP might interact with RNA elements within the HIV promoters. However, neither deletion of the Tat activation response (TAR) element (downstream of −20) of the HIV-1 promoter (Fig. 7A) nor mutation of the region between +1 and +20 (not shown) altered the effect of GLI-2/THP on promoter-driven expression. The experiments shown in Fig. 7 were performed in Jurkat T cells; very similar results were obtained in the U937 monocytic cell line (not shown). Therefore, in contrast to HTLV-1, the effect of GLI-2/THP on HIV gene expression does not appear to be mediated by known upstream (or downstream) enhancer elements.
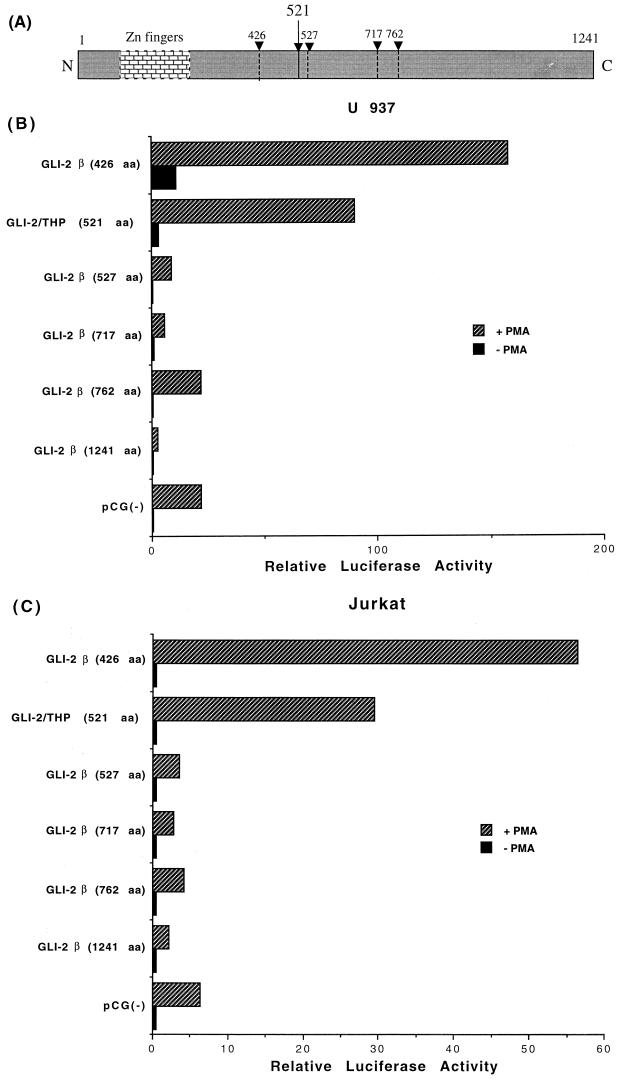
Only truncated forms of GLI-2 modulate retroviral transcription.
An individual GLI protein is typically found in multiple different forms in the cell, and variants which are less abundant are often biologically very significant (4, 75). GLI-2/THP is an isoform found in low abundance which is consistently seen at the RNA level (Fig. 6) but is seen consistently at the protein level only following purification steps (9a). As several new, larger isoforms of human GLI-2 have been described (85), we tested whether the longest (1,241 aa) GLI-2 construct, GLI-2β, would activate HIV-1 transcription. As seen in Fig. 8, this larger isoform did not increase HIV transcription in either T cells or monocytic cells, in contrast to the THP isoform. Thus, deletion mutants of GLI-2β were constructed and tested for transcriptional activity. The shortest of these mutants, GLI-2β (426 aa), which is 82 aa shorter than THP (508 aa), was the only one able to activate HIV-1 transcription (Fig. 8), perhaps yet more potently than THP. The longer deletion constructs had little effect on gene expression. Similar findings were noted with full-length and truncated versions of mouse GLI-2 (data not shown). Full-length (1,241 aa) GLI-2β, unlike any of the mutant constructs, was able to consistently, although only modestly, suppress expression from the HIV-1 promoter in both cell types (Fig. 8).
FIG. 8.
Only the truncated form of GLI-2β activates HIV-1 expression. (A) Representation of deletion mutants of human GLI-2β (dashed lines) relative to full-length (1,241 aa) and GLI-2/THP (521 aa). (B) U937 cells were transfected with 5 μg of HIV-1 reporter plasmid and 10 μg of empty vector or GLI-2-expressing constructs. Half of the cells were treated with 16 nM PMA after 24 h. Cells were lysed 48 h posttransfection, and the extracts were assayed for luciferase activity. (C) Jurkat cells were transfected and analyzed as for panel B.
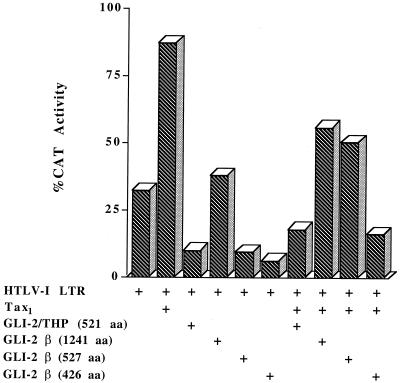
We then tested the ability of the same mutant constructs of GLI-2 to suppress expression from the HTLV-1 promoter. Again, GLI-2β (426 aa) had the most potent effect, followed by THP (Fig. 9). Unlike the case for HIV-1, GLI-2β (527 aa) also showed some effect. Full-length GLI-2β (1241 aa) had little effect in the presence of Tax and no significant effect in the absence of Tax (Fig. 9). Therefore, while there were some differences, just as in the HIV-1 system, it is the shorter forms of GLI-2 which modulate gene expression. Thus, GLI-2 shows similarities and differences to cubitus interruptus. Like cubitus interruptus, a small, less abundant protein form is involved in transcriptional repression. However, the long form of cubitus interruptus is the one which activates gene expression, whereas for GLI-2/THP the truncated form not only represses but also activates gene expression, at least with these retroviral promoters. The long isoforms of human GLI-2 would thus appear to contain a domain which inhibits their ability to modulate transcription.
FIG. 9.
Ability of GLI-2 constructs to suppress expression from the HTLV-1 LTR. Transient transfections of CV-1 cells were performed with 7.5 μg of a plasmid containing the full-length HTLV-1 LTR upstream of a CAT reporter gene (pU3R-CAT) in the presence or absence of 0.5 μg of a plasmid expressing Tax1 and 4 μg of a plasmid expressing GLI-2/THP, GLI-2β (1,241 aa), GLI-2β (527 aa), or GLI-2β (426 aa) as indicated. Forty-eight hours after transfection, extracts were prepared and analyzed by CAT assay using [14C]chloramphenicol and thin-layer chromatography. The results were analyzed with a PhosphorImager and ImageQuant software. The extent of transactivation of the HTLV-1 LTR is represented as percent CAT activity.
GLI-2/THP stimulation of HIV-1 replication is dependent on the first two zinc fingers.
The ability of GLI-2/THP to activate HIV promoter-driven transcription suggested that this form of human GLI-2 might be able to stimulate HIV replication. Further, the reporter gene transfection experiments suggested that the first, and possibly second, zinc finger might be necessary for this effect, if seen (Fig. 5). We thus tested the ability of GLI-2/THP, or constructs in which the first, second, or first and second zinc fingers had been mutated at a single amino acid, to stimulate HIV-1 replication. We found that GLI-2/THP can markedly stimulate HIV-1 replication in freshly transfected U937 monocytic cells (Fig. 10A and reference 9a) and in the chronically infected U1 monocytic cell line (Fig. 10B). Mutation of either zinc finger 1 or 2 markedly reduced the ability of GLI-2/THP to stimulate viral replication. Thus, GLI-2/THP can stimulate both early and latent viral replication, and does so through a mechanism which involves zinc fingers which are minimally involved in DNA binding (Fig. 5A and reference 63).
DISCUSSION
In the present report, the following key findings are presented. (i) GLI-2/THP, an isoform of GLI-2, is a potent modulator of retroviral transcription and replication. This observation links the vital GLI proteins to the replication of human pathogenic retroviruses. Thus, this work identifies a new function for the GLI family and also a new mechanism by which retroviral gene expression and replication might be controlled in cells. (ii) While GLI-2/THP was originally called the Tax helper protein, this protein unexpectedly suppresses HTLV transcription rather than activating it. Its ability to suppress HTLV-1 transcription and activate HIV-1 and HIV-2 transcription is reminiscent of a related protein, YY1, which is also noted to be capable of both activating and repressing transcription. (iii) The mechanism by which GLI-2/THP works to modulate retroviral transcription is different than expected. In the case of HTLV-1, merely mutating the minimal pets site does not alter suppression by this transcription factor. However, deletion of a larger region which includes the pets site does eliminate suppression. Thus, it would appear that GLI-2 is part of a complex of cellular proteins and enhancer elements which likely work together. In the case of HIV-1 and HIV-2, GLI-2/THP does not work through any obvious upstream (or downstream) enhancer element, thus demonstrating that not all modulation of transcription by GLI proteins need proceed through TG-rich enhancer elements. (iv) The first and second zinc fingers of GLI-2/THP, which are not important for DNA binding, are vital to both activation and suppression of gene expression, a finding which runs somewhat counter to prevailing thought (see below). Further, it is the small isoform of GLI-2, THP, which is active in both transcriptional activation and repression, again dissimilar to observations made with other GLI proteins, such as cubitus interruptus.
Our findings in the above studies using naturally occurring human retroviral promoters lead to different conclusions than do the studies of others (85, 86). These authors concluded that the larger isoforms of human GLI-2 were the biologically significant ones, based on their inability to detect the THP form of GLI-2 in certain cell lines. However, they were able to detect an RNA message compatible with the THP form of GLI-2 (86). Our observations also suggest that on the protein level, THP is not an abundant form of human GLI-2, and purification steps must be used in order to consistently identify this isoform of the protein in cellular extracts (9a). However, as noted above, it is not unusual for less abundant isoforms of GLI proteins to be biologically very significant, as is the case for the small form of cubitus interruptus (4, 75). These observations, as well as the potency of the GLI-2/THP effect on retroviral transcription and replication, strongly suggest that human GLI-2/THP is a biologically significant form of human GLI-2. This conclusion is also supported by the observation that mouse GLI-2/THP, but not full-length GLI-2, is also a potent activator of transcription (M. Smith and D. Markovitz, unpublished observations).
Another difference between our findings and those reported elsewhere concerns the question of what the functional effect of GLI-2/THP is on Tax-mediated activation of the HTLV-1 promoter. It has been suggested that GLI-2 is a transcriptional activator which synergizes with Tax to stimulate HTLV-1. However, previous studies involved overexpression of a larger form of human GLI-2 (GLI-2α) and employed CAT reporter plasmids which had copies of individual Tax-responsive sequences rather than the naturally occurring HTLV-1 promoter used in our studies. Even in those experiments, overexpression of human GLI-2α actually suppressed CAT expression, unless GLI-2 was linked to a GAL4 protein/reporter system (85). It was also demonstrated that GLI-2 can interact with CREB, which binds to the 21-bp repeats found next to the TG-rich (pets) element which GLI-2 can bind (20). However, no functional demonstration that CREB and GLI-2 can act synergistically was presented.
As discussed above, GLI family members are widely conserved in nature, being found to be important in the development of nematodes (tra-1 [21, 32, 61, 95, 96]), zebrafish (45), Drosophila (cubitus interruptus [3, 23, 42]), Xenopus (53), mice (40, 56, 90, 92), and humans (GLI-1, GLI-2/THP, and GLI-3 [44, 47, 73, 76, 77, 89]) and in human disease (18, 47, 76, 77). While human GLI proteins have been shown to bind to TG-rich promoter elements, in some cases in functionally significant sites (14, 85, 86), and can modulate transcription in constructs in which multiple GLI binding sites are placed upstream of a heterologous promoter (1, 53, 78), the present study is one of the few (19) to report on a human GLI protein able to modulate expression driven by natural promoters. Similar to the related YY-1 protein, GLI-2/THP is able to both suppress (HTLV-1) and activate (HIV-1 and HIV-2) gene expression, while having no apparent effect on other related promoters (HTLV-2). Interestingly, GLI-2/THP function requires an intact first zinc finger, which is not necessary to bind DNA, but not a VP16-like domain or a CREB binding protein (CBP) binding domain, both of which have been observed to be crucial for the ability of GLI-3 and GLI-1 to activate transcription (19, 93). GLI-2/THP generally requires the presence of PMA to activate HIV replication and gene expression. As PMA does not stimulate the expression of GLI-2/THP itself, it appears that PMA must either stimulate the expression of, or modify, a GLI-2/THP cofactor, decrease the expression or function of a GLI-2/THP suppressor, or directly induce a posttranslational effect on GLI-2/THP itself. This is reminiscent of cubitus interruptus, which appears to affect transcription not simply on the basis of the expressed level of the protein, which stays constant, but following posttranslational modifications (such as proteolysis [4]) or protein-protein interactions. The function of cubitus interruptus is also modulated by a protein kinase, with its effect being diminished by exposure to protein kinase A (11; reviewed in reference 65). It has also been shown that cubitus interruptus is found in a complex with the serine-threonine kinase Fused (72).
Regulation of HTLV-1 transcription by GLI-2/THP, while not depending simply on the TG-rich minimally defined pets site as we suspected, does require the region of the promoter which includes the pets element. The regulation of HIV-2 transcription by GLI-2/THP is, surprisingly, not mediated through the TG-rich pets enhancer element to which this protein binds in vitro. This suggests that the binding of other GLI proteins, such as GLI-1, to similar sites may also not always hold the key to understanding how these important proteins function to regulate gene expression. Regulation of a specific, naturally occurring promoter by a given GLI protein, demonstrated to be mediated by specific GLI binding sites, has only very recently been reported (19). While previous data suggest that cubitus interruptus can act through GLI binding sites to modulate transcription (3), direct evidence showing activation of gene expression (driven by the wingless promoter) mediated by GLI binding sites has only recently been presented (88). Even in the latter study, the effect of cubitus interruptus on the intact promoter was minimal (<2-fold stimulation), and only by placing an isolated, very distal fragment of the wingless promoter in front of a heterologous promoter was sufficient activation obtained to perform a site-specific mutational analysis confirming the importance of the specific binding sites. Therefore, our report is one of the first to present data relevant to the mechanism by which a GLI protein activates expression from intact, naturally occurring promoters.
In Drosophila, only one true GLI protein, cubitus interruptus, is known to exist, whereas humans and mice have at least three GLI proteins. It has been suggested that whereas cubitus interruptus serves as both an activator and repressor of gene expression, the three known human GLI proteins have evolved specialized functions, each serving as either an activator or repressor of transcription and each competing with the other for GLI binding sites (74). Our findings do not favor this model, as GLI-2/THP can clearly either repress or activate gene expression, depending on the specific target promoter and the state of activation of signal transduction pathways. Further, our data demonstrate that not all modulation of gene expression by GLI proteins takes place through the TG-rich binding sites predicted by in vitro DNA binding studies. A second theory which suggests that different domains of the individual GLI proteins might separately mediate activation and repression has been presented (7, 19). Our data also do not support this model, as mutation of the first zinc finger of GLI-2/THP, which does not alter DNA binding, markedly affects the ability of GLI-2/THP to both activate and repress gene expression. Thus, like cubitus interruptus and GLI-3, GLI-2 can induce both transcriptional repression and activation. However, unlike the situation with these proteins, the same domain, the first zinc finger, is involved in both forms of regulation. In our experiments, cells generally had to be treated with PMA to demonstrate activation of the HIV-1 and HIV-2 promoters by GLI-2/THP. Therefore, it appears likely that, as is the case for cubitus interruptus, phosphorylation changes can modulate the function of human GLI proteins. Further study of how GLI-2 modulates transcription should shed light both on the mechanism of action of the developmentally important GLI proteins and on retroviral gene expression and replication, a process which appears to exploit these highly conserved proteins.
ACKNOWLEDGMENTS
We thank C.-C. Hui for helpful comments and Mitsuaki Yoshida for the gift of GLI-2 expression plasmids. U1 cells (from Tom Folks) and p89.6 (from Ron Collman) were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH.
This work was supported by grants AI36685 and AI30924 from the NIH to D.M.M. N.M.C. was supported by grant K08-AI01293 from the NIH and by an Infectious Diseases Society of America Young Investigator Award. C.M.B. was supported by the Cellular Biotechnology Training Program (5 T32 GM08353) and the Cancer Biology Training Program (T32 CA09676) of the University of Michigan. B.R.L. was supported by the Medical Scientist Training Program (NIGMS T32 GM07863) of the University of Michigan and the Harvey Fellows Program. S.D.G. was supported by a Research Associate Award from the Department of Veterans Affairs.
REFERENCES
- 1.Akimaru H, Chen Y, Dai P, Hou D-X, Nonaka M, Smolik S M, Armstrong S, Goodman R H, Ishii S. Drosophila CBP is a co-activator of cubitus interruptus in hedgehog signalling. Nature. 1997;386:735–738. doi: 10.1038/386735a0. [DOI] [PubMed] [Google Scholar]
- 2.Aldovini A, Walker B D. Techniques in HIV research. New York, N.Y: Stockton Press; 1990. [Google Scholar]
- 3.Alexandre C, Jacinto A, Ingham P W. Transcriptional activation of hedgehog target genes in Drosophila is mediated directly by the Cubitus interruptus protein, a member of the GLI family of zinc finger DNA-binding proteins. Genes Dev. 1996;10:2003–2013. doi: 10.1101/gad.10.16.2003. [DOI] [PubMed] [Google Scholar]
- 4.Aza-Blanc P, Ramirez-Weber F-A, Laget M-P, Schwartz C, Kornberg T B. Proteolysis that is inhibited by hedgehog targets cubitus interruptus protein to the nucleus and converts it to a repressor. Cell. 1997;89:1043–1053. doi: 10.1016/s0092-8674(00)80292-5. [DOI] [PubMed] [Google Scholar]
- 5.Barre-Sinoussi F, Chermann J C, Rey F, Nugeyre M T, Chamaret S, Gruest J, et al. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS) Science. 1983;220:868–871. doi: 10.1126/science.6189183. [DOI] [PubMed] [Google Scholar]
- 6.Bielinska A, Krasnow S, Nabel G J. NF-κB-mediated activation of the human immunodeficiency virus enhancer: site of transcriptional initiation is independent of the TATA box. J Virol. 1989;63:4097–4100. doi: 10.1128/jvi.63.9.4097-4100.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biesecker L G. Strike three for GLI3. Nat Genet. 1997;17:259–260. doi: 10.1038/ng1197-259. [DOI] [PubMed] [Google Scholar]
- 8.Bosselut R, Duvall J F, Gegonne A, Bailly M, Hemar A, Brady J, Ghysdael J. The product of the c-ets-1 proto-oncogene and the related Ets2 protein act as transcriptional activators of the long terminal repeat of human T cell leukemia virus HTLV-I. EMBO J. 1990;9:3137–3144. doi: 10.1002/j.1460-2075.1990.tb07511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Browning C, Hilfinger J M, Rainier S, Lin V, Hedderwick S, Smith M, Markovitz D M. The sequence and structure of the 3′ arm of the first stem-loop of the human immunodeficiency virus type 2 trans-activation responsive region mediate Tat-2 transactivation. J Virol. 1997;71:8048–8055. doi: 10.1128/jvi.71.10.8048-8055.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9a.Browning C M, Smith M J, Clark N M, Lane B R, Parada C, Montano M, KewalRamani V N, Littman D R, Essex M, Roeder R G, Markovitz D M. Human GLI-2 is a Tat activation response element-independent Tat cofactor. J Virol. 2001;75:2314–2323. doi: 10.1128/JVI.75.5.2314-2323.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen I S Y, Cann A J, Shah N P, Gaynor R B. Functional relation between HTLV-II x and adenovirus E1A proteins in transcriptional activation. Science. 1985;230:570–573. doi: 10.1126/science.2996140. [DOI] [PubMed] [Google Scholar]
- 11.Chen Y, Cardinaux J-R, Goodman R H, Smolik S M. Mutants of cubitus interruptus that are independent of PKA regulation are independent of hedgehog signaling. Development. 1999;126:3607–3616. doi: 10.1242/dev.126.16.3607. [DOI] [PubMed] [Google Scholar]
- 12.Chen Y, Struhl G. Dual roles for Patched in sequestering and transducing Hedgehog. Cell. 1996;87:553–563. doi: 10.1016/s0092-8674(00)81374-4. [DOI] [PubMed] [Google Scholar]
- 13.Clark N M, Hannibal M C, Markovitz D M. The peri-κB site of the human immunodeficiency virus type 2 enhancer is active in monocytes but not T cells. J Virol. 1995;69:4854–4862. doi: 10.1128/jvi.69.8.4854-4862.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clark N M, Smith M J, Hilfinger J M, Markovitz D M. Activation of the human T-cell leukemia virus type I enhancer is mediated by binding sites for Elf-1 and the pets factor. J Virol. 1993;67:5522–5528. doi: 10.1128/jvi.67.9.5522-5528.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clavel F, Guetard D, Brun-Vezinet F, Chamaret S, Rey M A, Santos-Ferreira O, et al. Isolation of a new human retrovirus from West African patients with AIDS. Science. 1986;233:343–346. doi: 10.1126/science.2425430. [DOI] [PubMed] [Google Scholar]
- 16.Clavel F, Guyader M, Guetard D, Salle M, Montagnier L, Alizon M. Molecular cloning and polymorphism of the human immune deficiency virus type 2. Nature. 1986;324:691–695. doi: 10.1038/324691a0. [DOI] [PubMed] [Google Scholar]
- 17.Clavel F, Mansinho K, Chamaret S, Guetard D, Favier V, Nina J, et al. Human immunodeficiency virus type 2 infection associated with AIDS in West Africa. N Engl J Med. 1987;316:1180–1185. doi: 10.1056/NEJM198705073161903. [DOI] [PubMed] [Google Scholar]
- 18.Dahmane N, Lee J, Robins P, Heller P, Ruiz i Altaba A. Activation of the transcription factor Gli1 and the Sonic hedgehog signalling pathway in skin tumours. Nature. 1997;389:876–881. doi: 10.1038/39918. [DOI] [PubMed] [Google Scholar]
- 19.Dai P, Akimaru H, Tanaka Y, Maekawa T, Nakafuku M, Ishii S. Sonic hedgehog-induced activation of the Gli1 promoter is mediated by GLI3. J Biol Chem. 1999;274:8143–8152. doi: 10.1074/jbc.274.12.8143. [DOI] [PubMed] [Google Scholar]
- 20.Dan S, Tanimura A, Yoshida M. Interaction of Gli2 with CREB protein on DNA elements in the long terminal repeat of human T-cell leukemia virus type 1 is responsible for transcriptional activation by Tax protein. J Virol. 1999;73:3258–3263. doi: 10.1128/jvi.73.4.3258-3263.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Bono M, Zarkower D, Hodgkin J. Dominant feminizing mutations implicate protein-protein interactions as the main mode of regulation of the nematode sex-determining gene tra-1. Genes Dev. 1995;9:155–167. doi: 10.1101/gad.9.2.155. [DOI] [PubMed] [Google Scholar]
- 22.Ding Q, Motoyama J, Gasca S, Mo R, Sasaki H, Rossant J, Hui C-C. Diminished sonic hedgehog signaling and lack of floor plate differentiation in Gli2 mutant mice. Development. 1998;125:2533–2543. doi: 10.1242/dev.125.14.2533. [DOI] [PubMed] [Google Scholar]
- 23.Dominguez M, Brunner M, Hafen E, Basler K. Sending and receiving the hedgehog signal: control by the Drosophila Gli protein cubitus interruptus. Science. 1996;272:1621–1625. doi: 10.1126/science.272.5268.1621. [DOI] [PubMed] [Google Scholar]
- 24.Fu G, Grosveld G, Markovitz D M. DEK, an autoantigen involved in a chromosomal translocation in acute myelogenous leukemia, binds to the human immunodeficiency virus type 2 enhancer. Proc Natl Acad Sci USA. 1997;94:1811–1815. doi: 10.1073/pnas.94.5.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fu G K, Markovitz D M. Purification of the pets factor: a nuclear protein that binds to the inducible TG-rich element of the human immunodeficiency virus type 2 enhancer. J Biol Chem. 1996;271:19599–19605. doi: 10.1074/jbc.271.32.19599. [DOI] [PubMed] [Google Scholar]
- 26.Fu G K, Smith M J, Markovitz D M. Bacterial protease Lon is a site-specific DNA-binding protein. J Biol Chem. 1997;272:534–538. [PubMed] [Google Scholar]
- 27.Gallo R C, Salahuddin S Z, Popovic M, Shearer G M, Kaplan M, Haynes B F, et al. Frequent detection and isolation of cytopathic retroviruses (HTLV-III) from patients with AIDS and at risk for AIDS. Science. 1984;224:500–503. doi: 10.1126/science.6200936. [DOI] [PubMed] [Google Scholar]
- 28.Gegonne A, Bosselut R, Bailly R-A, Ghysdael J. Synergistic activation of the HTLV-I LTR ets-responsive region by transcription factors ets-1 and Sp1. EMBO J. 1993;12:1169–1178. doi: 10.1002/j.1460-2075.1993.tb05758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gessain A, Vernant J C, Maurs L, Barin F, Gout O, Calendar A, de The G. Antibodies to human T-lymphotropic virus type-I in patients with tropical spastic paraparesis. Lancet. 1985;ii:407–410. doi: 10.1016/s0140-6736(85)92734-5. [DOI] [PubMed] [Google Scholar]
- 30.Gitlin S D, Bosselut R, Gegonne A, Ghysdael J, Brady J N. Sequence-specific interaction of the Ets1 protein with the long terminal repeat of human T-lymphotropic virus type I. J Virol. 1991;65:5513–5523. doi: 10.1128/jvi.65.10.5513-5523.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gorman C M, Moffat L F, Howard B H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Graves L E, Segal S, Goodwin E B. TRA-1 regulates the cellular distribution of the tra-2 mRNA in C. elegans. Nature. 1999;399:802–805. doi: 10.1038/21682. [DOI] [PubMed] [Google Scholar]
- 33.Guyader M, Emerman M, Sonigo P, Clavel F, Montagnier L, Alizon M. Genome organization and transactivation of the human immunodeficiency virus type 2. Nature. 1987;326:662–669. doi: 10.1038/326662a0. [DOI] [PubMed] [Google Scholar]
- 34.Hahn B H, Shaw G M, Arya S K, Popovic M, Gallo R C, Wong-Staal F. Molecular cloning and characterization of HTLV-III virus associated with AIDS. Nature. 1984;312:166–169. doi: 10.1038/312166a0. [DOI] [PubMed] [Google Scholar]
- 35.Hahn H, Wicking C, Zaphiropoulos P G, Gailani M R, Shanley S, Chidambaram A, Vorechovsky I, Holmberg E, Unden A B, Gillies S, Negus K, Smyth I, Pressman C, Leffell D J, Gerrard B, Goldstein A M, Dean M, Toftgard R, Chenevix-Trench G, Wainwright B, Bale A E. Mutations of the human homolog of Drosophila patched in the nevoid basal cell carcinoma syndrome. Cell. 1996;85:841–851. doi: 10.1016/s0092-8674(00)81268-4. [DOI] [PubMed] [Google Scholar]
- 36.Hannibal M C, Markovitz D M, Clark N, Nabel G J. Differential activation of human immunodeficiency virus type 1 and 2 transcription by specific T-cell activation signals. J Virol. 1993;67:5035–5040. doi: 10.1128/jvi.67.8.5035-5040.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hannibal M C, Markovitz D M, Nabel G J. Multiple cis-acting elements in the human immunodeficiency virus type 2 enhancer mediate the response to T-cell receptor stimulation by antigen in a T-cell hybridoma line. Blood. 1994;83:1839–1846. [PubMed] [Google Scholar]
- 38.Hardcastle Z, Mo R, Hui C-C, Sharpe P T. The Shh signalling pathway in tooth development: defects in Gli2 and Gli3 mutants. Development. 1998;125:2803–2811. doi: 10.1242/dev.125.15.2803. [DOI] [PubMed] [Google Scholar]
- 39.Hilfinger J, Clark N, Robinson K, Smith M, Markovitz D M. Differential regulation of the human immunodeficiency virus type 2 in monocytic cells at varying stages of differentiation. J Virol. 1993;67:4448–4453. doi: 10.1128/jvi.67.7.4448-4453.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hui C-C, Slusarski D, Platt K A, Holmgren R, Joyner A L. Expression of three mouse homologs of the Drosophila segment polarity gene cubitus interruptus, GLI, GLI-2, and GLI-3, in ectoderm- and mesoderm-derived tissues suggests multiple roles during postimplantation development. Dev Biol. 1994;162:402–413. doi: 10.1006/dbio.1994.1097. [DOI] [PubMed] [Google Scholar]
- 41.Jeang K-T, Boros I, Brady J, Radonovich M, Khoury G. Characterization of cellular factors that interact with the human T-cell leukemia virus type I p40x-responsive 21-base-pair sequence. J Virol. 1988;62:4499–4509. doi: 10.1128/jvi.62.12.4499-4509.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johnson R L, Rothman A L, Xie J, Goodrich L V, Bare J W, Bonifas J M, Quinn A G, Myers R M, Cox D R, Epstein E H, Jr, Scott M P. Human homolog of patched, a candidate gene for the basal cell nevus syndrome. Science. 1996;272:1668–1671. doi: 10.1126/science.272.5268.1668. [DOI] [PubMed] [Google Scholar]
- 43.Kalyanaraman V S, Sarngadharan M G, Robert-Guroff M, Miyoshi I, Blayne D, Golde D, et al. A new subtype of human T-cell leukemia virus (HTLV-II) associated with a T-cell variant of hairy cell leukemia. Science. 1982;218:571–573. doi: 10.1126/science.6981847. [DOI] [PubMed] [Google Scholar]
- 44.Kang S, Graham J M, Jr, Olney A H, Biesecker L G. GLI3 frameshift mutations cause autosomal dominant Pallister-Hall syndrome. Nat Genet. 1997;15:266–268. doi: 10.1038/ng0397-266. [DOI] [PubMed] [Google Scholar]
- 45.Karlstrom R O, Talbot W S, Schier A F. Comparative synteny cloning of zebrafish you-too: mutations in the Hedgehog target gli2 affect ventral forebrain patterning. Genes Dev. 1999;13:388–393. doi: 10.1101/gad.13.4.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kinoshita S, Lishan S, Amano M, Timmerman L A, Kaneshima H, Nolan G P. The T cell activation factor NF-ATc positively regulates HIV-1 replication and gene expression in T cells. Immunity. 1997;6:235–244. doi: 10.1016/s1074-7613(00)80326-x. [DOI] [PubMed] [Google Scholar]
- 47.Kinzler K W, Bigner S H, Bigner D D, Trent J M, Law M L, O'Brien S J, Wong A J, Vogelstein B. Identification of an amplified, highly expressed gene in a human glioma. Science. 1987;236:70–73. doi: 10.1126/science.3563490. [DOI] [PubMed] [Google Scholar]
- 48.Leiden J M, Wang C-W, Petryniak B, Markovitz D M, Nabel G J, Thompson C B. A novel ets-related transcription factor, Elf-1, binds to the human immunodeficiency virus type 2 regulatory elements that are required for inducible trans-activation in T cells. J Virol. 1992;66:5890–5897. doi: 10.1128/jvi.66.10.5890-5897.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leung K, Nabel G J. HTLV-I transactivator induces interleukin-2 receptor expression through an NF-κB-like factor. Nature. 1988;333:776–778. doi: 10.1038/333776a0. [DOI] [PubMed] [Google Scholar]
- 50.Licht J D, Grossel M J, Figge J, Hansen U M. Drosophila Krüppel protein is a transcriptional repressor. Nature. 1990;346:76–79. doi: 10.1038/346076a0. [DOI] [PubMed] [Google Scholar]
- 51.Margolis D M, Somasundaran M, Green M R. Human transcription factor YY1 represses human immunodeficiency virus type 1 transcription and virion production. J Virol. 1994;68:905–910. doi: 10.1128/jvi.68.2.905-910.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marigo V, Davey R A, Zuo Y, Cunningham J M, Tabin C J. Biochemical evidence that Patched is the Hedgehog receptor. Nature. 1996;384:176–179. doi: 10.1038/384176a0. [DOI] [PubMed] [Google Scholar]
- 53.Marine J-C, Bellefroid E J, Pendeville H, Martial J A, Pieler T. A role for Xenopus Gli-type zinc finger proteins in the early embryonic patterning of mesoderm and neuroectoderm. Mech Dev. 1997;63:211–225. doi: 10.1016/s0925-4773(97)00050-6. [DOI] [PubMed] [Google Scholar]
- 54.Markovitz D M, Hannibal M, Perez V L, Gauntt C, Folks T M, Nabel G J. Differential regulation of human immunodeficiency viruses (HIVs): a specific regulatory element in HIV-2 responds to stimulation of the T cell antigen receptor. Proc Natl Acad Sci USA. 1990;87:9098–9102. doi: 10.1073/pnas.87.23.9098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Markovitz D M, Smith M J, Hilfinger J, Hannibal M C, Petryniak B, Nabel G J. Activation of the human immunodeficiency virus type 2 enhancer is dependent on purine box and κB regulatory elements. J Virol. 1992;66:5479–5484. doi: 10.1128/jvi.66.9.5479-5484.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mo R, Freer A M, Zinyk D L, Crackower M A, Michaud J, Heng H H-Q, Chik K W, Shi X-M, Tsui L-C, Cheng S H, Joyner A L, Hui C-C. Specific and redundant functions of Gli2 and Gli3 zinc finger genes in skeletal patterning and development. Development. 1997;124:113–123. doi: 10.1242/dev.124.1.113. [DOI] [PubMed] [Google Scholar]
- 57.Motoyama J, Liu J, Mo R, Ding Q, Post M, Hui C-C. Essential function of Gli2 and Gli3 in the formation of lung, trachea and oesophagus. Nat Genet. 1998;20:54–57. doi: 10.1038/1711. [DOI] [PubMed] [Google Scholar]
- 58.Nabel G, Baltimore D. An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature. 1987;326:711–713. doi: 10.1038/326711a0. [DOI] [PubMed] [Google Scholar]
- 59.Nabel G J, Rice S A, Knipe D M, Baltimore D. Alternative mechanisms for activation of human immunodeficiency virus enhancer in T cells. Science. 1988;23:1299–1302. doi: 10.1126/science.2830675. [DOI] [PubMed] [Google Scholar]
- 60.Niki M, Ohtani K, Nakamura M, Sugamura K. Multistep regulation of enhancer activity of the 21-base-pair element of human T-cell leukemia virus type I. J Virol. 1992;66:4348–4357. doi: 10.1128/jvi.66.7.4348-4357.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Orenic T V, Slusarski D C, Kroll K L, Holmgren R A. Cloning and characterization of the segment polarity gene cubitus interruptus Dominant of Drosophila. Genes Dev. 1990;4:1053–1067. doi: 10.1101/gad.4.6.1053. [DOI] [PubMed] [Google Scholar]
- 62.Osame M, Usuku K, Izumo S, Ijichi N, Amitani H, Igata A, Matsumoto M, Tara M. HTLV-I associated myelopathy, a new clinical entity. Lancet. 1986;i:1031–1032. doi: 10.1016/s0140-6736(86)91298-5. [DOI] [PubMed] [Google Scholar]
- 63.Pavletich N P, Pabo C O. Crystal structure of a five-finger GLI-DNA complex: new perspectives on zinc fingers. Science. 1993;261:1701–1707. doi: 10.1126/science.8378770. [DOI] [PubMed] [Google Scholar]
- 64.Perkins N D, Edwards N L, Duckett C S, Agranoff A B, Schmidt R M, Nabel G J. A cooperative interaction between NF-κB and Sp1 is required for HIV-1 enhancer activation. EMBO J. 1993;12:3551–3558. doi: 10.1002/j.1460-2075.1993.tb06029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Perrimon N. Hedgehog and beyond. Cell. 1995;80:517–520. doi: 10.1016/0092-8674(95)90503-0. [DOI] [PubMed] [Google Scholar]
- 66.Poiesz B J, Ruscetti F W, Gazadar A F, Bunn P A, Minna J D, Gallo R C. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc Natl Acad Sci USA. 1980;77:7415–7419. doi: 10.1073/pnas.77.12.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Poiesz B J, Ruscetti F W, Reitz M S, Kalyanaraman V S, Gallo R C. Isolation of a new type C retrovirus (HTLV) in primary uncultured cells of a patient with Sezary T-cell leukaemia. Nature. 1981;294:268–271. doi: 10.1038/294268a0. [DOI] [PubMed] [Google Scholar]
- 68.Popovic M, Sarngadharan M G, Read E, Gallo R C. Detection, isolation, and continuous production of cytopathic retroviruses (HTLV-III) from patients with AIDS and pre-AIDS. Science. 1984;224:497–500. doi: 10.1126/science.6200935. [DOI] [PubMed] [Google Scholar]
- 69.Poteat H T, Chen F Y, Kadison P, Sodroski J G, Haseltine W A. Protein kinase A-dependent binding of a nuclear factor to the 21-base-pair repeat of the human T-cell leukemia virus type I long terminal repeat. J Virol. 1990;64:1264–1270. doi: 10.1128/jvi.64.3.1264-1270.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Queen C, Baltimore D. Immunoglobulin gene transcription is activated by downstream sequence elements. Cell. 1983;33:741–748. doi: 10.1016/0092-8674(83)90016-8. [DOI] [PubMed] [Google Scholar]
- 71.Radhakrishna U, Wild A, Grzeschik K-H, Antonarakis S E. Mutation in GLI3 in postaxial polydactyly type A. Nat Genet. 1997;17:269–271. doi: 10.1038/ng1197-269. [DOI] [PubMed] [Google Scholar]
- 72.Robbins D J, Nybakken K E, Kobayashi R, Sisson J C, Bishop J M, Therond P P. Hedgehog elicits signal transduction by means of a large complex containing the kinesin-related protein Costal 2. Cell. 1997;90:225–234. doi: 10.1016/s0092-8674(00)80331-1. [DOI] [PubMed] [Google Scholar]
- 73.Roberts M, Douglass E C, Peiper S C, Houghton P J, Look A T. Amplification of the gli gene in childhood sarcomas. Cancer Res. 1989;49:5407–5413. [PubMed] [Google Scholar]
- 74.Ruiz i Altaba A. Catching a Gli-mpse of Hedgehog. Cell. 1997;90:193–196. doi: 10.1016/s0092-8674(00)80325-6. [DOI] [PubMed] [Google Scholar]
- 75.Ruiz i Altaba A. Gli proteins encode context-dependent positive and negative functions: implications for development and disease. Development. 1999;126:3205–3216. doi: 10.1242/dev.126.14.3205. [DOI] [PubMed] [Google Scholar]
- 76.Ruppert J M, Kinzler K W, Wong A J, Bigner S H, Kao F-T, Law M L, Seuanez H N, O'Brien S J, Vogelstein B. The GLI-Krüppel family of human genes. Mol Cell Biol. 1988;8:3104–3113. doi: 10.1128/mcb.8.8.3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ruppert J M, Vogelstein B, Kinzler K W. The zinc finger protein GLI transforms primary cells in cooperation with adenovirus E1A. Mol Cell Biol. 1991;11:1724–1728. doi: 10.1128/mcb.11.3.1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sasaki H, Hui C-C, Nakafuku M, Kondoh H. A binding site for Gli proteins is essential for HNF-3β floor plate enhancer activity in transgenics and can respond to Shh in vitro. Development. 1997;124:1313–1322. doi: 10.1242/dev.124.7.1313. [DOI] [PubMed] [Google Scholar]
- 79.Seiki M, Hattori S, Hirayama Y, Yoshida M. Human adult T-cell leukemia virus: complete nucleotide sequence of the provirus genome integrated in leukemia cell DNA. Proc Natl Acad Sci USA. 1983;80:3618–3622. doi: 10.1073/pnas.80.12.3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shi Y, Seto E, Chang L-S, Shenk T. Transcriptional repression by YY1, a human GLI-Krüppel-related protein, and relief of repression by adenovirus E1A protein. Cell. 1991;67:377–388. doi: 10.1016/0092-8674(91)90189-6. [DOI] [PubMed] [Google Scholar]
- 81.Shin S H, Kogerman P, Lindstrom E, Tofgard R, Biesecker L G. GLI3 mutations in human disorders mimic Drosophila cubitus interruptus protein functions and localization. Proc Natl Acad Sci USA. 1999;96:2880–2884. doi: 10.1073/pnas.96.6.2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Smith M R, Greene W C. Molecular biology of the type I human T-cell leukemia virus (HTLV-I) and adult T-cell leukemia. J Clin Investig. 1991;87:761–766. doi: 10.1172/JCI115078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sodroski J G, Rosen C A, Haseltine W A. trans-acting transcriptional activation of the long terminal repeat of human T lymphotropic viruses in infected cells. Science. 1984;225:381–385. doi: 10.1126/science.6330891. [DOI] [PubMed] [Google Scholar]
- 84.Stone D M, Hynes M, Armanini M, Swanson T A, Gu Q, Johnson R L, Scott M P, Pennica D, Goddard A, Phillips H, Noll M, Hooper J E, de Sauvage F, Rosenthal A. The tumour-suppressor gene patched encodes a candidate receptor for Sonic hedgehog. Nature. 1996;384:129–134. doi: 10.1038/384129a0. [DOI] [PubMed] [Google Scholar]
- 85.Tanimura A, Dan S, Yoshida M. Cloning of novel isoforms of the human Gli2 oncogene and their activities to enhance tax-dependent transcription of the human T-cell leukemia virus type 1 genome. J Virol. 1998;72:3958–3964. doi: 10.1128/jvi.72.5.3958-3964.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tanimura A, Teshima H, Fujisawa J-I, Yoshida M. A new regulatory element that augments the Tax-dependent enhancer of human T-cell leukemia virus type 1 and cloning of cDNAs encoding its binding proteins. J Virol. 1993;67:5375–5382. doi: 10.1128/jvi.67.9.5375-5382.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Usheva A, Shenk T. TATA-binding protein-independent initiation: YY1, TFIIB, and RNA polymerase II direct basal transcription on supercoiled template DNA. Cell. 1994;76:1115–1121. doi: 10.1016/0092-8674(94)90387-5. [DOI] [PubMed] [Google Scholar]
- 88.Von Ohlen T, Lessing D, Nusse R, Hooper J E. Hedgehog signaling regulates transcription through cubitus interruptus, a sequence-specific DNA binding protein. Proc Natl Acad Sci USA. 1997;94:2404–2409. doi: 10.1073/pnas.94.6.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vortkamp A, Gessler M, Grzeschik K-H. GLI3 zinc-finger gene interrupted by translocations in Greig syndrome families. Nature. 1991;352:539–540. doi: 10.1038/352539a0. [DOI] [PubMed] [Google Scholar]
- 90.Walterhouse D, Ahmed M, Slusarski D, Kalamaras J, Boucher D, Holmgren R, Iannaccone P. gli, a zinc finger transcription factor and oncogene, is expressed during normal mouse development. Dev Dyn. 1993;196:91–102. doi: 10.1002/aja.1001960203. [DOI] [PubMed] [Google Scholar]
- 91.Xiao H, Goldthwait D A, Mapstone T. A search for Gli expression in tumors of the central nervous system. Pediatr Neurosurg. 1994;20:178–182. doi: 10.1159/000120783. [DOI] [PubMed] [Google Scholar]
- 92.Yang J T, Liu C Z, Villavicencio E H, Yoon J W, Walterhouse D, Iannaccone P M. Expression of human GLI in mice results in failure to thrive, early death, and patchy Hirschsprung-like gastrointestinal dilatation. Mol Med. 1997;3:826–835. [PMC free article] [PubMed] [Google Scholar]
- 93.Yoon J W, Liu C Z, Yang J T, Swart R, Iannaccone P, Walterhouse D. GLI activates transcription through a herpes simplex viral protein 16-like activation domain. J Biol Chem. 1998;273:3496–3501. doi: 10.1074/jbc.273.6.3496. [DOI] [PubMed] [Google Scholar]
- 94.Yoshida M, Miyoshi I, Hinuma Y. Isolation and characterization of retrovirus from cell lines of human adult T-cell leukemia and its implication in the disease. Proc Natl Acad Sci USA. 1982;79:2031–2035. doi: 10.1073/pnas.79.6.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zarkower D, Hodgkin J. Molecular analysis of the C. elegans sex-determining gene tra-1: a gene encoding two zinc finger proteins. Cell. 1992;70:237–249. doi: 10.1016/0092-8674(92)90099-x. [DOI] [PubMed] [Google Scholar]
- 96.Zarkower D, Hodgkin J. Zinc fingers in sex determination: only one of the two C.elegans Tra-1 proteins binds DNA in vitro. Nucleic Acids Res. 1993;21:3691–3698. doi: 10.1093/nar/21.16.3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zuo P, Stanojevic D, Colgan J, Han K, Levine M, Manley J L. Activation and repression of transcription by the gap proteins hunchback and Krüppel in cultured Drosophila cells. Genes Dev. 1991;5:254–264. doi: 10.1101/gad.5.2.254. [DOI] [PubMed] [Google Scholar]