FIGURE 6:
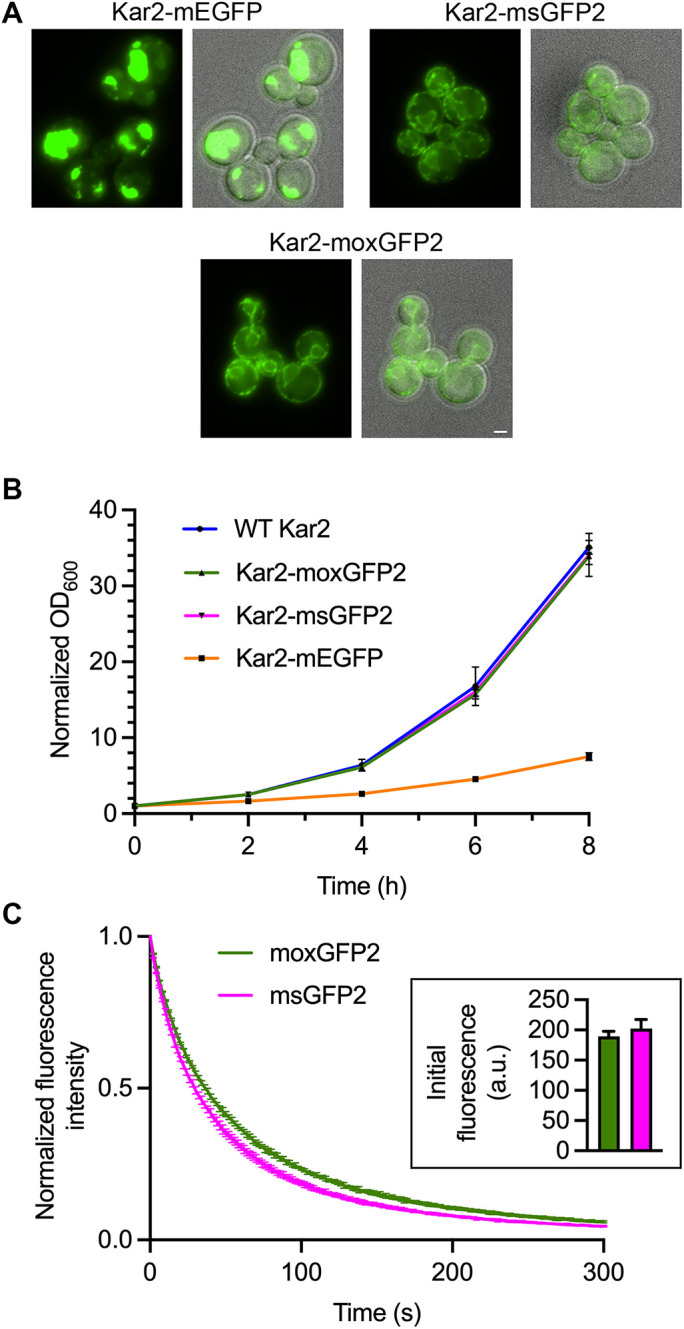
moxGFP2 is suitable for use as a fluorescent tag in the yeast ER lumen. (A) Fluorescence and merged fluorescence/brightfield images of yeast strains in which the endogenous KAR2 gene was replaced with the indicated GFP fusion gene. Scale bar, 2 μm. (B) Growth in rich liquid medium of yeast strains expressing either wild-type (“WT”) Kar2 or the indicated GFP fusions to Kar2. The optical densities at 600 nm (OD600) were ∼0.1 at the beginning of the experiment, and those initial values were normalized to 1.0. (C) Photostability of moxGFP2 and msGFP2 fused to Kar2 in live yeast cells. Confocal z-stacks were captured every 2 s for 5 min and then average projected to create movies. Plotted are averaged fluorescence signals from three movies per strain, where each movie included ∼10–15 cells. Mean fluorescence signals were normalized to the values at time zero. The bars represent SEM values. Shown in the inset are mean cellular fluorescence signals with SEM values for the first timepoints of the Kar2-moxGFP2 movies (green) and the Kar2-msGFP2 movies (magenta).
