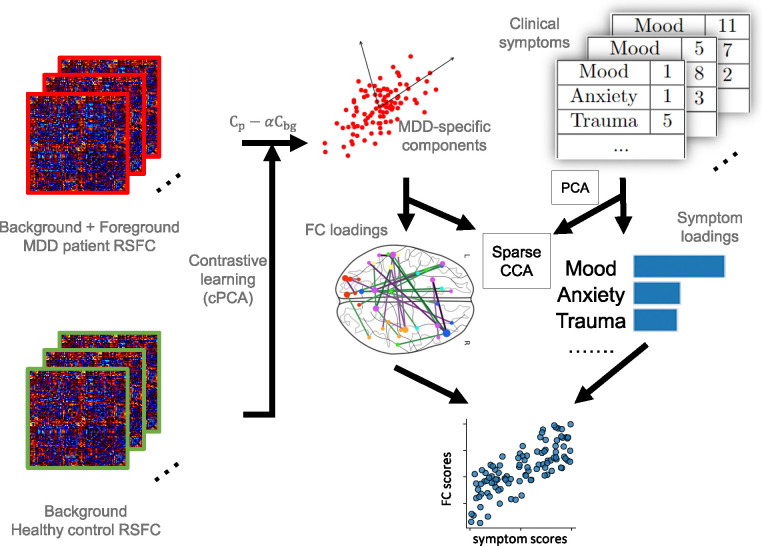
Figure 1.
Illustration of the proposed framework. MDD patient RSFC data is considered to include both disorder-specific information (foreground) and shared noise with healthy control data (background). Contrastive learning is used for removing the background noise from patient data to generate MDD-specific components. Specifically, the covariance matrix of patient RSFC () and healthy control RSFC () are computed to represent the group variances. MDD-specific components are derived from the updated covariance matrix , which represents the foreground variance, and represents a hyperparameter quantifying the degree of contrast. These components are used as MDD-specific RSFC features in the next step. Subsequently, sparse CCA is performed to identify the linked dimensions between neurophysiology and PCA-transformed symptoms, represented by FC and symptom loadings. These loadings serve as linear combinations which project the RSFC features and symptoms into a common space with high correlations.