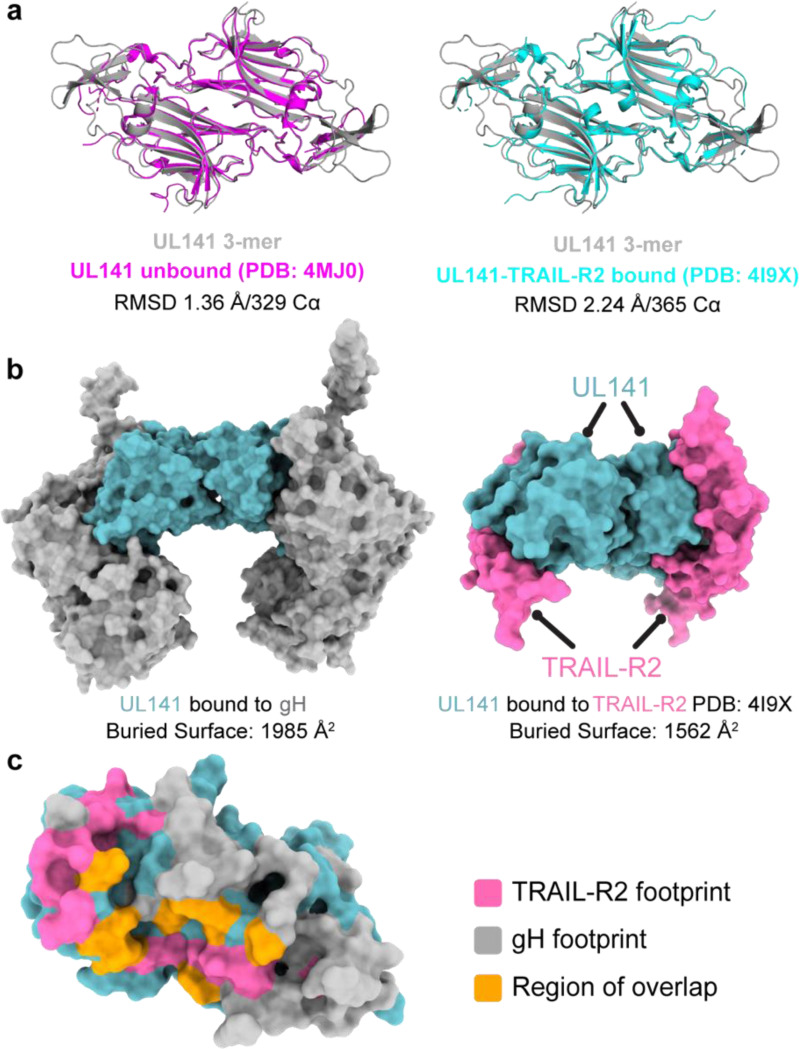
Fig. 13. gH and TRAIL-R2 share a similar binding site on UL141.
a, Structural comparison of UL141 from the 3-mer with unbound UL141 (left) and UL141 bound to TRAIL-R2 (right). The dimer structures were aligned as “dimers” using all Cα atoms in the respective PDB files, with the alignment quantified by the indicated r.m.s.d. values. b, Surface models of UL141 in the 3-mer (left) and UL141 bound to TRAIL-R2 (right) show that gH and TRAIL-R2 occupy similar binding sites on UL141. The calculated buried surface area for each interaction is indicated below. c, Surface model of a UL141 monomer with the TRAIL-R2 binding footprint highlighted in pink, the gH footprint in grey, and the region of overlap in orange. The calculated buried surface area of the overlapping region is indicated, accounting for approximately 25% of the TRAIL-R2 binding site.