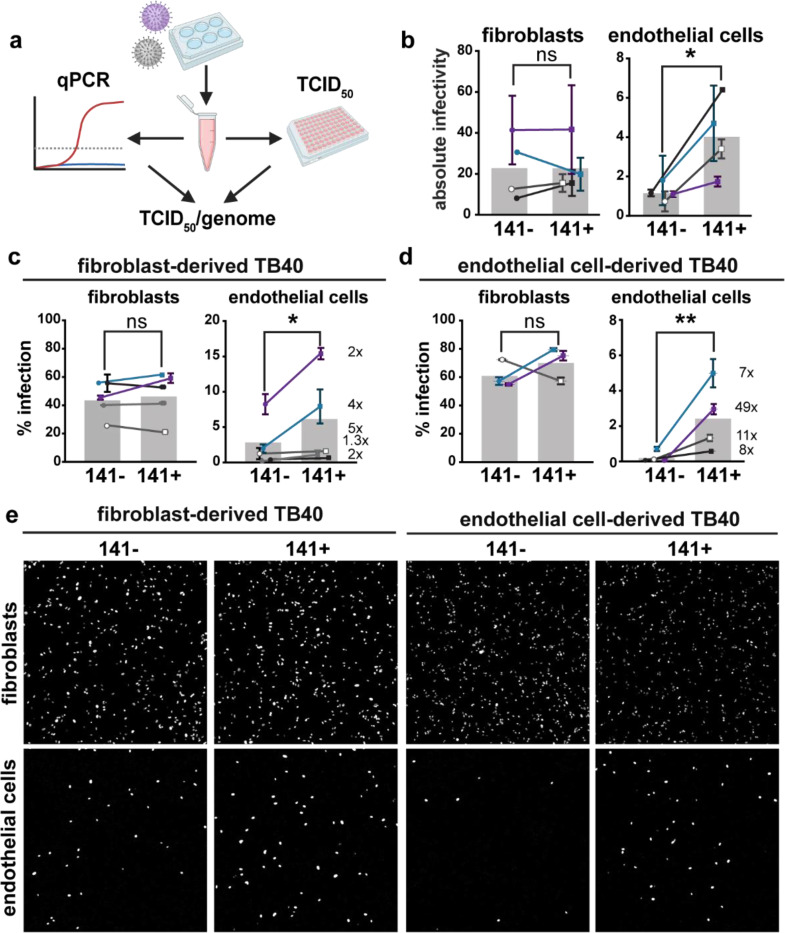
Fig. 2. The 3-mer improves endothelial cell infectivity.
a, Schematic of methods used to measure the absolute infectivity of TB40 virions. Viral genomes/mL were calculated from a standard curve with a 10-fold dilution series of TB40 BAC DNA. TCID50 assays were performed on fibroblasts and HUVEC in parallel by staining for IE1 at 3 days post infection, and together were used to calculate TCID50/genome (BioRender.com). b, Absolute infectivity shown as TCID50/103 genomes for fibroblasts and endothelial cells, and data represent 4 biological replicates. c, Infectivity of fibroblast-derived and d, endothelial cell-derived 141+ or 141− HCMV TB40 with fold differences shown as #x for each biological replicate. Fibroblasts and endothelial cells were infected for 24h in parallel with 50 or 100 genome equivalents/cell, respectively, stained for IE1 and counterstained with Hoechst 33342 to calculate the percentage of infected cells. e, Representative images of c and d. Statistical significance was determined by a paired t-test, with each point representing a biological replicate consisting of 2–3 technical replicates each. Error bars represent ± SEM for each biological replicate. Shaded bars are the mean % infection for all biological replicates.