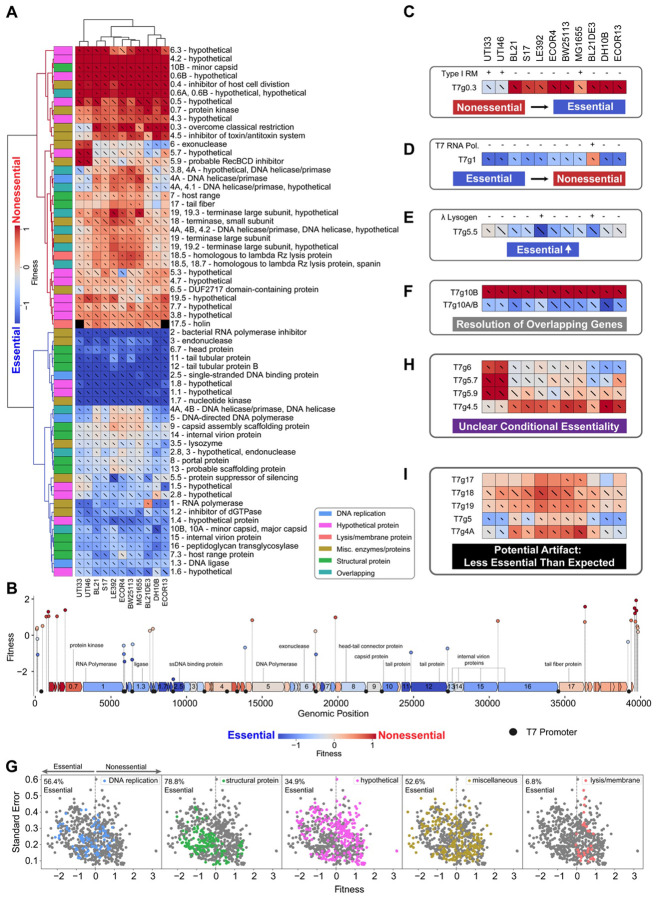
Figure 2. PhageMaP recapitulates established biology and uncovers novel insights in model T7 phage.
(A) Heatmap of the T7 fitness scores after Z-normalization within each of the 11 conditions. To better emphasize differences outside the two extremes of the data, a 10th percentile floor and 90th percentile ceiling were imposed for the color scaling. The lines through each cell represent the standard error (SE) where a full line represents SE≥1. Clustering was performed using the Euclidean metric and Ward linkage method. Metadata describes general classification of each gene. (B) Plot mapping the genomic coordinates and scores for intergenic perturbations. The points (intergenic variants) and blocks (genes) are colored using the same scale as (A). The gene scores used for coloring are the medians of scores from each tested condition. Instances from (A) of (C) generally nonessential genes that become more essential. The presence of intact Type I restriction-modification systems was determined by used PADLOC121. Instances from (A) of (D) generally essential genes that become more nonessential, (E) moderately essential gene becoming more essential, and (F) resolution of overlapping genetic elements. (G) Scatter plots showing the fitness scores and standard errors for each fitness measurement for the 11 hosts. Points corresponding to each classification are highlighted by color. Dotted lines represent the threshold for “nonessential” and “essential” genes. Instances from (A) of (H) unclear conditional essentiality and (I) potential artifacts involving characterized genes.