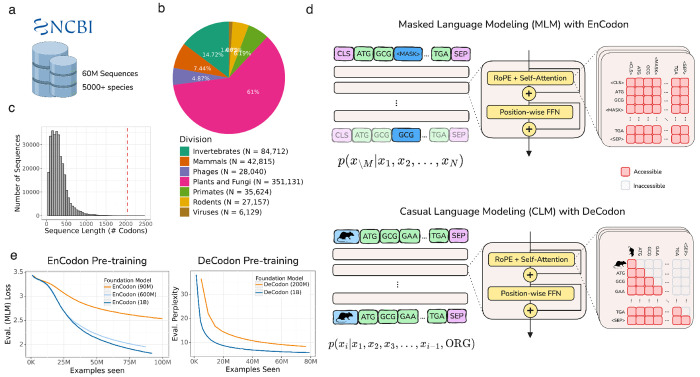
Figure 1: Overview of EnCodon and DeCodon:
a) Over 60 million coding sequences from 5000 species has been extracted from NCBI Genomes database and used to pre-train EnCodon and DeCodon foundation models. b) An overwhelming majority of the data (98.7%) is comprised of bacterial coding sequences. Pie chart depicting division makeup of non-bacterial coding sequences in NCBI is shown. c) Histogram of coding sequence lengths (number of codons) in NCBI Genomes database. We used 2048 as maximum sequence length supported by EnCodon and DeCodon based taking the shown distrbution into account to cover more than 99.8% of sequences. d) We pretrained EnCodon using masked language modeling (MLM) objective where parts of sequences were corrupted/masked and the model has to predict the true token at the positions given the rest of tokens (i.e. context). DeCodon is a conditional generative transformer model which provides controllable coding sequence generation by querying sequence organism as the very first input token. We pre-trained DeCodon with causal (autoregressive) language modeling objective on aggregated corpus of coding sequences where each sequence is prepended with a special organism token. Rotary Positional Self-Attention was used in both EnCodon and DeCodon blocks. e) 3 EnCodons and 2 DeCodons, differing in scale (i.e. number of trainable parameters) have been pre-trained for more than 1,000,000 optimization steps on the aggregated corpus from NCBI Genomes database.