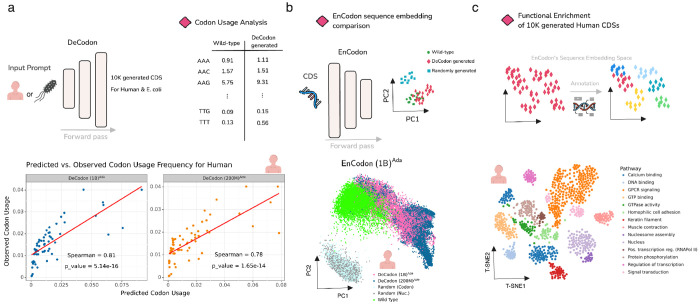
Figure 3: DeCodon generates functional and organism-specific coding sequences:
a) DeCodon takes organism as input and generates a coding sequence specific to the queried species. We generated 10,000 coding sequences (CDS) for Human and E. coli species. Scatter plots of codon usage frequencies of wild-type (y-axis) and generated (x-axis) is shown for human annotated with spearman correlation and associated p-value. b) To further compare the generated CDS population with the wild-type, we generated two groups of randomly sampled CDSs and computed sequence embeddings of wild-type, DeCodon generated, and randomly generated groups. PCA visualization of sequence embeddings is shown for human-related coding sequences. c) Finally, we used protein functional annotation tools to test the functional enrichment of the sequence clusters in EnCodon embedding space. We used InterProScan to predict functional domains of human-generated CDSs by DeCodon (1B)Ada. T-SNE visualization of functionally annotated generated sequences by DeCodon (1B)Ada is shown where generated sequences were colored by their enriched biological pathway.